Novel long noncoding RNAs (lncRNAs) in myogenesis: a miR-31 overlapping lncRNA transcript controls myoblast differentiation
- PMID: 25512605
- PMCID: PMC4301723
- DOI: 10.1128/MCB.01394-14
Novel long noncoding RNAs (lncRNAs) in myogenesis: a miR-31 overlapping lncRNA transcript controls myoblast differentiation
Erratum in
-
Correction for Ballarino et al., "Novel Long Noncoding RNAs (lncRNAs) in Myogenesis: a miR-31 Overlapping lncRNA Transcript Controls Myoblast Differentiation".Mol Cell Biol. 2018 May 29;38(12):e00167-18. doi: 10.1128/MCB.00167-18. Print 2018 Jun 15. Mol Cell Biol. 2018. PMID: 29844107 Free PMC article. No abstract available.
Abstract
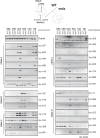
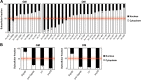
Transcriptome analysis allowed the identification of new long noncoding RNAs differentially expressed during murine myoblast differentiation. These transcripts were classified on the basis of their expression under proliferating versus differentiated conditions, muscle-restricted activation, and subcellular localization. Several species displayed preferential expression in dystrophic (mdx) versus wild-type muscles, indicating their possible link with regenerative processes. One of the identified transcripts, lnc-31, even if originating from the same nuclear precursor of miR-31, is produced by a pathway mutually exclusive. We show that lnc-31 and its human homologue hsa-lnc-31 are expressed in proliferating myoblasts, where they counteract differentiation. In line with this, both species are more abundant in mdx muscles and in human Duchenne muscular dystrophy (DMD) myoblasts, than in their normal counterparts. Altogether, these data suggest a crucial role for lnc-31 in controlling the differentiation commitment of precursor myoblasts and indicate that its function is maintained in evolution despite the poor sequence conservation with the human counterpart.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
