A posttranscriptional mechanism that controls Ptbp1 abundance in the Xenopus epidermis
- PMID: 25512611
- PMCID: PMC4301716
- DOI: 10.1128/MCB.01040-14
A posttranscriptional mechanism that controls Ptbp1 abundance in the Xenopus epidermis
Abstract
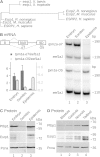
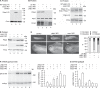
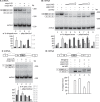
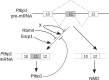
The output of alternative splicing depends on the cooperative or antagonistic activities of several RNA-binding proteins (RBPs), like Ptbp1 and Esrp1 in Xenopus. Fine-tuning of the RBP abundance is therefore of prime importance to achieve tissue- or cell-specific splicing patterns. Here, we addressed the mechanisms leading to the high expression of the ptbp1 gene, which encodes Ptbp1, in Xenopus epidermis. Two splice isoforms of ptbp1 mRNA differ by the presence of an alternative exon 11, and only the isoform including exon 11 can be translated to a full-length protein. In vivo minigene assays revealed that the nonproductive isoform was predominantly produced. Knockdown experiments demonstrated that Esrp1, which is specific to the epidermis, strongly stimulated the expression of ptbp1 by favoring the productive isoform. Consequently, knocking down esrp1 phenocopied ptbp1 inactivation. Conversely, Ptbp1 repressed the expression of its own gene by favoring the nonproductive isoform. Hence, a complex posttranscriptional mechanism controls Ptbp1 abundance in Xenopus epidermis: skipping of exon 11 is the default splicing pattern, but Esrp1 stimulates ptbp1 expression by favoring the inclusion of exon 11 up to a level that is limited by Ptbp1 itself. These results decipher a posttranscriptional mechanism that achieves various abundances of the ubiquitous RBP Ptbp1 in different tissues.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Robust identification of Ptbp1-dependent splicing events by a junction-centric approach in Xenopus laevis.Dev Biol. 2017 Jun 15;426(2):449-459. doi: 10.1016/j.ydbio.2016.08.021. Epub 2016 Aug 19. Dev Biol. 2017. PMID: 27546377
-
Regulation of mRNA abundance by polypyrimidine tract-binding protein-controlled alternate 5' splice site choice.PLoS Genet. 2014 Nov 6;10(11):e1004771. doi: 10.1371/journal.pgen.1004771. eCollection 2014 Nov. PLoS Genet. 2014. PMID: 25375251 Free PMC article.
-
High expression of PTBP1 promote invasion of colorectal cancer by alternative splicing of cortactin.Oncotarget. 2017 May 30;8(22):36185-36202. doi: 10.18632/oncotarget.15873. Oncotarget. 2017. PMID: 28404950 Free PMC article.
-
The role of RNA splicing factor PTBP1 in neuronal development.Biochim Biophys Acta Mol Cell Res. 2023 Oct;1870(7):119506. doi: 10.1016/j.bbamcr.2023.119506. Epub 2023 May 31. Biochim Biophys Acta Mol Cell Res. 2023. PMID: 37263298 Review.
-
The Emerging Role of the RBM20 and PTBP1 Ribonucleoproteins in Heart Development and Cardiovascular Diseases.Genes (Basel). 2020 Apr 8;11(4):402. doi: 10.3390/genes11040402. Genes (Basel). 2020. PMID: 32276354 Free PMC article. Review.
Cited by
-
RAVER1 hinders lethal EMT and modulates miR/RISC activity by the control of alternative splicing.Nucleic Acids Res. 2024 Apr 24;52(7):3971-3988. doi: 10.1093/nar/gkae046. Nucleic Acids Res. 2024. PMID: 38300787 Free PMC article.
-
Roles of PTBP1 in alternative splicing, glycolysis, and oncogensis.J Zhejiang Univ Sci B. 2020 Feb.;21(2):122-136. doi: 10.1631/jzus.B1900422. Epub 2020 Feb 5. J Zhejiang Univ Sci B. 2020. PMID: 32115910 Free PMC article. Review.
-
Perspective in Alternative Splicing Coupled to Nonsense-Mediated mRNA Decay.Int J Mol Sci. 2020 Dec 10;21(24):9424. doi: 10.3390/ijms21249424. Int J Mol Sci. 2020. PMID: 33321981 Free PMC article. Review.
-
Polypyrimidine Tract Binding Protein: A Universal Player in Cancer Development.Curr Mol Med. 2024;24(12):1450-1460. doi: 10.2174/0115665240251370231017053236. Curr Mol Med. 2024. PMID: 37877563 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
