ITS1, 5.8S and ITS2 secondary structure modelling for intra-specific differentiation among species of the Colletotrichum gloeosporioides sensu lato species complex
- PMID: 25512885
- PMCID: PMC4254888
- DOI: 10.1186/2193-1801-3-684
ITS1, 5.8S and ITS2 secondary structure modelling for intra-specific differentiation among species of the Colletotrichum gloeosporioides sensu lato species complex
Abstract
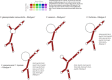
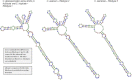
The Colletotrichum gloeosporioides species complex is among the most destructive fungal plant pathogens in the world, however, identification of member species which are of quarantine importance is impacted by a number of factors that negatively affect species identification. Structural information of the rRNA marker may be considered to be a conserved marker which can be used as supplementary information for possible species identification. The difficulty in using ITS rDNA sequences for identification lies in the low level of sequence variation at the intra-specific level and the generation of artificially-induced sequence variation due to errors in polymerization of the ITS array during DNA replication. Type and query ITS sequences were subjected to sequence analyses prior to generation of predicted consensus secondary structures, including the pattern of nucleotide polymorphisms and number of indel haplotypes, GC content, and detection of artificially-induced sequence variation. Data pertaining to structure stability, the presence of conserved motifs in secondary structures and mapping of all sequences onto the consensus C. gloeosporioides sensu stricto secondary structure for ITS1, 5.8S and ITS2 markers was then carried out. Motifs that are evolutionarily conserved among eukaryotes were found for all ITS1, 5.8S and ITS2 sequences. The sequences exhibited conserved features typical of functional rRNAs. Generally, polymorphisms occurred within less conserved regions and were seen as bulges, internal and terminal loops or non-canonical G-U base-pairs within regions of the double stranded helices. Importantly, there were also taxonomic motifs and base changes that were unique to specific taxa and which may be used to support intra-specific identification of members of the C. gloeosporioides sensu lato species complex.
Keywords: Colletotrichum spp; Internally transcribed spacer region; Secondary structure prediction.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

