Foxc1 dependent mesenchymal signalling drives embryonic cerebellar growth
- PMID: 25513817
- PMCID: PMC4281880
- DOI: 10.7554/eLife.03962
Foxc1 dependent mesenchymal signalling drives embryonic cerebellar growth
Abstract
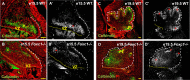
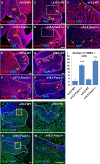
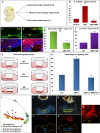
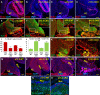
Loss of Foxc1 is associated with Dandy-Walker malformation, the most common human cerebellar malformation characterized by cerebellar hypoplasia and an enlarged posterior fossa and fourth ventricle. Although expressed in the mouse posterior fossa mesenchyme, loss of Foxc1 non-autonomously induces a rapid and devastating decrease in embryonic cerebellar ventricular zone radial glial proliferation and concurrent increase in cerebellar neuronal differentiation. Subsequent migration of cerebellar neurons is disrupted, associated with disordered radial glial morphology. In vitro, SDF1α, a direct Foxc1 target also expressed in the head mesenchyme, acts as a cerebellar radial glial mitogen and a chemoattractant for nascent Purkinje cells. Its receptor, Cxcr4, is expressed in cerebellar radial glial cells and conditional Cxcr4 ablation with Nes-Cre mimics the Foxc1-/- cerebellar phenotype. SDF1α also rescues the Foxc1-/- phenotype. Our data emphasizes that the head mesenchyme exerts a considerable influence on early embryonic brain development and its disruption contributes to neurodevelopmental disorders in humans.
Keywords: Cxcl12; cerebellum; developmental biology; foxc1; mouse; neurodevelopmental disorder; neuroscience; radial glia; stem cells.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures





References
-
- Aldinger KA, Lehmann OJ, Hudgins L, Chizhikov VV, Bassuk AG, Ades LC, Krantz ID, Dobyns WB, Millen KJ. FOXC1 is required for normal cerebellar development and is a major contributor to chromosome 6p25.3 Dandy-Walker malformation. Nature Genetics. 2009;41:1037–1042. doi: 10.1038/ng.422. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

