Bacteria-bacteria interactions within the microbiota of the ancestral metazoan Hydra contribute to fungal resistance
- PMID: 25514534
- PMCID: PMC4478695
- DOI: 10.1038/ismej.2014.239
Bacteria-bacteria interactions within the microbiota of the ancestral metazoan Hydra contribute to fungal resistance
Abstract
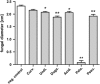
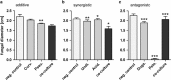
Epithelial surfaces of most animals are colonized by diverse microbial communities. Although it is generally agreed that commensal bacteria can serve beneficial functions, the processes involved are poorly understood. Here we report that in the basal metazoan Hydra, ectodermal epithelial cells are covered with a multilayered glycocalyx that provides a habitat for a distinctive microbial community. Removing this epithelial microbiota results in lethal infection by the filamentous fungus Fusarium sp. Restoring the complex microbiota in gnotobiotic polyps prevents pathogen infection. Although mono-associations with distinct members of the microbiota fail to provide full protection, additive and synergistic interactions of commensal bacteria are contributing to full fungal resistance. Our results highlight the importance of resident microbiota diversity as a protective factor against pathogen infections. Besides revealing insights into the in vivo function of commensal microbes in Hydra, our findings indicate that interactions among commensal bacteria are essential to inhibit pathogen infection.
Figures



References
-
- Augustin R, Anton-Erxleben F, Jungnickel S, Hemmrich G, Spudy B, Podschun R, et al. Activity of the novel peptide arminin against multiresistant human pathogens shows the considerable potential of phylogenetically ancient organisms as drug sources. Antimicrob Agents Chemother. 2009;53:5245–5250. - PMC - PubMed
-
- Augustin R, Bosch TCG. Cnidarian immunity: a tale of two barriers. Adv Exp Med Biol. 2010;708:1–16. - PubMed
-
- Augustin R, Siebert S, Bosch TCG. Identification of a kazal-type serine protease inhibitor with potent anti-staphylococcal activity as part of Hydra's innate immune system. Dev Comp Immunol. 2009;33:830–837. - PubMed
-
- Bohnhoff M, Drake BL, Miller CP. The effect of an antibiotic on the susceptibility of the mouse's intestinal tract to Salmonella infection. Antibiot Annu. 1955;3:453–455. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

