Single molecule analysis of functionally asymmetric G protein-coupled receptor (GPCR) oligomers reveals diverse spatial and structural assemblies
- PMID: 25516594
- PMCID: PMC4326798
- DOI: 10.1074/jbc.M114.622498
Single molecule analysis of functionally asymmetric G protein-coupled receptor (GPCR) oligomers reveals diverse spatial and structural assemblies
Abstract
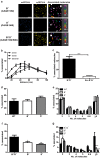
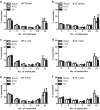
Formation of G protein-coupled receptors (GPCRs) into dimers and higher order oligomers represents a key mechanism in pleiotropic signaling, yet how individual protomers function within oligomers remains poorly understood. We present a super-resolution imaging approach, resolving single GPCR molecules to ∼ 8 nm resolution in functional asymmetric dimers and oligomers using dual-color photoactivatable dyes and localization microscopy (PD-PALM). PD-PALM of two functionally defined mutant luteinizing hormone receptors (LHRs), a ligand-binding deficient receptor (LHR(B-)) and a signaling-deficient (LHR(S-)) receptor, which only function via intermolecular cooperation, favored oligomeric over dimeric formation. PD-PALM imaging of trimers and tetramers revealed specific spatial organizations of individual protomers in complexes where the ratiometric composition of LHR(B-) to LHR(S-) modulated ligand-induced signal sensitivity. Structural modeling of asymmetric LHR oligomers strongly aligned with PD-PALM-imaged spatial arrangements, identifying multiple possible helix interfaces mediating inter-protomer associations. Our findings reveal that diverse spatial and structural assemblies mediating GPCR oligomerization may acutely fine-tune the cellular signaling profile.
Keywords: Bioluminescence Resonance Energy Transfer (BRET); Cell Signaling; Dimer; G Protein; G protein-Coupled Receptor (GPCR); Oligomer; Structural Biology; Super-resolution Imaging.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures







References
-
- Pascal G., Milligan G. (2005) Functional complementation and the analysis of opioid receptor homodimerization. Mol. Pharmacol. 68, 905–915 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

