Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation
- PMID: 25517091
- PMCID: PMC6795551
- DOI: 10.1038/nature13994
Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation
Abstract
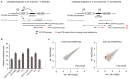
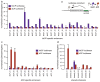
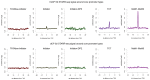
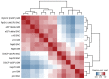
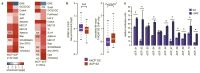
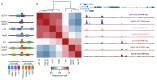
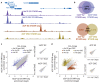
Gene transcription in animals involves the assembly of RNA polymerase II at core promoters and its cell-type-specific activation by enhancers that can be located more distally. However, how ubiquitous expression of housekeeping genes is achieved has been less clear. In particular, it is unknown whether ubiquitously active enhancers exist and how developmental and housekeeping gene regulation is separated. An attractive hypothesis is that different core promoters might exhibit an intrinsic specificity to certain enhancers. This is conceivable, as various core promoter sequence elements are differentially distributed between genes of different functions, including elements that are predominantly found at either developmentally regulated or at housekeeping genes. Here we show that thousands of enhancers in Drosophila melanogaster S2 and ovarian somatic cells (OSCs) exhibit a marked specificity to one of two core promoters--one derived from a ubiquitously expressed ribosomal protein gene and another from a developmentally regulated transcription factor--and confirm the existence of these two classes for five additional core promoters from genes with diverse functions. Housekeeping enhancers are active across the two cell types, while developmental enhancers exhibit strong cell-type specificity. Both enhancer classes differ in their genomic distribution, the functions of neighbouring genes, and the core promoter elements of these neighbouring genes. In addition, we identify two transcription factors--Dref and Trl--that bind and activate housekeeping versus developmental enhancers, respectively. Our results provide evidence for a sequence-encoded enhancer-core-promoter specificity that separates developmental and housekeeping gene regulatory programs for thousands of enhancers and their target genes across the entire genome.
Conflict of interest statement
The authors declare no competing financial interests.
Figures








Comment in
-
Enhancer trafficking: free throws and three-pointers.Dev Cell. 2015 Jan 26;32(2):135-7. doi: 10.1016/j.devcel.2015.01.006. Dev Cell. 2015. PMID: 25625204
-
Enhancers: holding out for the right promoter.Curr Biol. 2015 Mar 30;25(7):R290-3. doi: 10.1016/j.cub.2015.01.039. Curr Biol. 2015. PMID: 25829016 Free PMC article.
References
-
- Merli C, Bergstrom DE, Cygan JA, Blackman RK. Promoter specificity mediates the independent regulation of neighboring genes. Genes Dev. 1996;10:1260–1270. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

