Ferrets exclusively synthesize Neu5Ac and express naturally humanized influenza A virus receptors
- PMID: 25517696
- PMCID: PMC4351649
- DOI: 10.1038/ncomms6750
Ferrets exclusively synthesize Neu5Ac and express naturally humanized influenza A virus receptors
Abstract
Mammals express the sialic acids N-acetylneuraminic acid (Neu5Ac) and N-glycolylneuraminic acid (Neu5Gc) on cell surfaces, where they act as receptors for pathogens, including influenza A virus (IAV). Neu5Gc is synthesized from Neu5Ac by the enzyme cytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMAH). In humans, this enzyme is inactive and only Neu5Ac is produced. Ferrets are susceptible to human-adapted IAV strains and have been the dominant animal model for IAV studies. Here we show that ferrets, like humans, do not synthesize Neu5Gc. Genomic analysis reveals an ancient, nine-exon deletion in the ferret CMAH gene that is shared by the Pinnipedia and Musteloidia members of the Carnivora. Interactions between two human strains of IAV with the sialyllactose receptor (sialic acid--α2,6Gal) confirm that the type of terminal sialic acid contributes significantly to IAV receptor specificity. Our results indicate that exclusive expression of Neu5Ac contributes to the susceptibility of ferrets to human-adapted IAV strains.
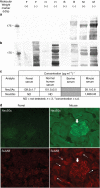
Figures

References
-
- Patterson K. D. & Pyle G. F. The geography and mortality of the 1918 influenza pandemic. Bull. Hist. Med. 65, 4–21 (1991). - PubMed
-
- Rogers G. N. et al. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature 304, 76–78 (1983). - PubMed
-
- Herlocher M. L. et al. Assessment of development of resistance to antivirals in the ferret model of influenza virus infection. J. Infect. Dis. 188, 1355–1361 (2003). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

