ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma
- PMID: 25517749
- PMCID: PMC4269829
- DOI: 10.1016/j.ccell.2014.09.019
ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma
Abstract
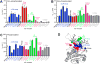
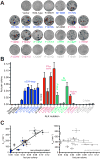
Genetic studies have established anaplastic lymphoma kinase (ALK), a cell surface receptor tyrosine kinase, as a tractable molecular target in neuroblastoma. We describe comprehensive genomic, biochemical, and computational analyses of ALK mutations across 1,596 diagnostic neuroblastoma samples. ALK tyrosine kinase domain mutations occurred in 8% of samples--at three hot spots and 13 minor sites--and correlated significantly with poorer survival in high- and intermediate-risk neuroblastoma. Biochemical and computational studies distinguished oncogenic (constitutively activating) from nononcogenic mutations and allowed robust computational prediction of their effects. The mutated variants also showed differential in vitro crizotinib sensitivities. Our studies identify ALK genomic status as a clinically important therapeutic stratification tool in neuroblastoma and will allow tailoring of ALK-targeted therapy to specific mutations.
Copyright © 2014 Elsevier Inc. All rights reserved.
Conflict of interest statement
All other authors declare that they have no competing interests.
Figures


References
-
- Attiyeh EF, London WB, Mossé YP, Wang Q, Winter C, Khazi D, McGrady PW, Seeger RC, Look AT, Shimada H, et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–2253. - PubMed
-
- Bagci O, Tumer S, Olgun N, Altungoz O. Copy number status and mutation analyses of anaplastic lymphoma kinase (ALK) gene in 90 sporadic neuroblastoma tumors. Cancer Letts. 2012;317:72–77. - PubMed
-
- Bossi RT, Saccardo MB, Ardini E, Menichincheri M, Rusconi L, Magnaghi P, Orsini P, Avanzi N, Borgia AL, Nesi M, et al. Crystal structures of anaplastic lymphoma kinase in complex with ATP competitive inhibitors. Biochemistry. 2010;49:6813–6825. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

