Hepatitis C virus life cycle and lipid metabolism
- PMID: 25517881
- PMCID: PMC4280516
- DOI: 10.3390/biology3040892
Hepatitis C virus life cycle and lipid metabolism
Abstract
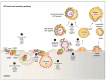
Hepatitis C Virus (HCV) infects over 150 million people worldwide. In most cases HCV infection becomes chronic, causing liver disease ranging from fibrosis to cirrhosis and hepatocellular carcinoma. HCV affects the cholesterol homeostasis and at the molecular level, every step of the virus life cycle is intimately connected to lipid metabolism. In this review, we present an update on the lipids and apolipoproteins that are involved in the HCV infectious cycle steps: entry, replication and assembly. Moreover, the result of the assembly process is a lipoviroparticle, which represents a peculiarity of hepatitis C virion. This review illustrates an example of an intricate virus-host interaction governed by lipid metabolism.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

