Evolution of the oxygen sensitivity of cytochrome c oxidase subunit 4
- PMID: 25519729
- PMCID: PMC4329467
- DOI: 10.1152/ajpregu.00281.2014
Evolution of the oxygen sensitivity of cytochrome c oxidase subunit 4
Abstract
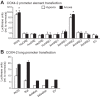
Vertebrates possess two paralogs of cytochrome c oxidase (COX) subunit 4: a ubiquitous COX4-1 and a hypoxia-linked COX4-2. Mammalian COX4-2 is thought to have a role in relation to fine-tuning metabolism in low oxygen levels, conferred through both structural differences in the subunit protein structure and regulatory differences in the gene. We sought to elucidate the pervasiveness of this feature across vertebrates. The ratio of COX4-2/4-1 mRNA is generally low in mammals, but this ratio was higher in fish and reptiles, particularly turtles. The COX4-2 gene appeared unresponsive to low oxygen in nonmammalian models (zebrafish, goldfish, tilapia, anoles, and turtles) and fish cell lines. Reporter genes constructed from the amphibian and reptile homologues of the mammalian oxygen-responsive elements and hypoxia-responsive elements did not respond to low oxygen. Unlike the rodent ortholog, the promoter of goldfish COX4-2 did not respond to hypoxia or anoxia. The protein sequences of the COX4-2 peptide showed that the disulfide bridge seen in human and rodent orthologs would be precluded in other mammalian lineages and lower vertebrates, all of which lack the requisite pair of cysteines. The coordinating ligands of the ATP-binding site are largely conserved across mammals and reptiles, but in Xenopus and fish, sequence variations may disrupt the ability of the protein to bind ATP at this site. Collectively, these results suggest that many of the genetic and structural features of COX4-2 that impart responsiveness and benefits in hypoxia may be restricted to the Euarchontoglires lineage that includes primates, lagomorphs, and rodents.
Keywords: COX4-2; HRE; ORE; oxygen sensitivity.
Copyright © 2015 the American Physiological Society.
Figures







References
-
- Arnold S. The power of life-cytochrome c oxidase takes center stage in metabolic control, cell signalling and survival. Mitochondrion 12: 46–56, 2012. - PubMed
-
- Arnold S, Kadenbach B. The intramitochondrial ATP/ADP-ratio controls cytochrome c oxidase activity allosterically. FEBS Lett 443: 105–108, 1999. - PubMed
-
- Bickler PE, Buck LT. Hypoxia tolerance in reptiles, amphibians, and fishes: life with variable oxygen availability. Annu Rev Physiol 69: 145–170, 2007. - PubMed
-
- Bremer K, Monk CT, Gurd BJ, Moyes CD. Transcriptional regulation of temperature-induced remodeling of muscle bioenergetics in goldfish. Am J Physiol Regul Integr Comp Physiol 303: R150–R158, 2012. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

