Targeting a cryptic allosteric site for selective inhibition of the oncogenic protein tyrosine phosphatase Shp2
- PMID: 25519989
- PMCID: PMC4303306
- DOI: 10.1021/bi5013595
Targeting a cryptic allosteric site for selective inhibition of the oncogenic protein tyrosine phosphatase Shp2
Abstract
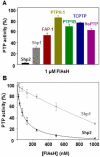
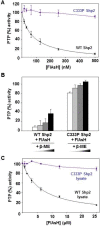
Protein tyrosine phosphatases (PTPs) have been the subject of considerable pharmaceutical-design efforts because of the ubiquitous connections between misregulation of PTP activity and human disease. PTP-inhibitor discovery has been hampered, however, by the difficulty in identifying cell-permeable compounds that can selectively target PTP active sites, and no PTP inhibitors have progressed to the clinic. The identification of allosteric sites on target PTPs therefore represents a potentially attractive solution to the druggability problem of PTPs. Here we report that the oncogenic PTP Shp2 contains an allosteric-inhibition site that renders the enzyme sensitive to potent and selective inhibition by cell-permeable biarsenical compounds. Because Shp2 contains no canonical tetracysteine biarsenical-binding motif, the enzyme's inhibitor-binding site is not readily predictable from its primary or three-dimensional structure. Intriguingly, however, Shp2's PTP domain does contain a cysteine residue (C333) at a position that is removed from the active site and is occupied by proline in other classical PTPs. We show that Shp2's unusual cysteine residue constitutes part of a Shp2-specific allosteric-inhibition site, and that Shp2's sensitivity to biarsenicals is dependent on the presence of the naturally occurring C333. The determinative role of this residue in conferring inhibitor sensitivity is surprising because C333's side chain is inaccessible to solvent in Shp2 crystal structures. The discovery of this cryptic Shp2 allosteric site may provide a means for targeting Shp2 activity with high specificity and suggests that buried-yet-targetable allosteric sites could be similarly uncovered in other protein families.
Figures




References
-
- Hendriks W. J.; Elson A.; Harroch S.; Pulido R.; Stoker A.; den Hertog J. (2013) Protein tyrosine phosphatases in health and disease. FEBS J. 280, 708–730. - PubMed
-
- Hardy S.; Julien S. G.; Tremblay M. L. (2012) Impact of oncogenic protein tyrosine phosphatases in cancer. Anti-Cancer Agents Med. Chem. 12, 4–18. - PubMed
-
- Chan G.; Kalaitzidis D.; Neel B. G. (2008) The tyrosine phosphatase Shp2 (PTPN11) in cancer. Cancer Metastasis Rev. 27, 179–192. - PubMed
-
- Mohi M. G.; Neel B. G. (2007) The role of Shp2 (PTPN11) in cancer. Curr. Opin. Genet. Dev. 17, 23–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

