Active destabilization of base pairs by a DNA glycosylase wedge initiates damage recognition
- PMID: 25520195
- PMCID: PMC4288190
- DOI: 10.1093/nar/gku1300
Active destabilization of base pairs by a DNA glycosylase wedge initiates damage recognition
Abstract
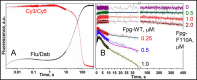
Formamidopyrimidine-DNA glycosylase (Fpg) excises 8-oxoguanine (oxoG) from DNA but ignores normal guanine. We combined molecular dynamics simulation and stopped-flow kinetics with fluorescence detection to track the events in the recognition of oxoG by Fpg and its mutants with a key phenylalanine residue, which intercalates next to the damaged base, changed to either alanine (F110A) or fluorescent reporter tryptophan (F110W). Guanine was sampled by Fpg, as evident from the F110W stopped-flow traces, but less extensively than oxoG. The wedgeless F110A enzyme could bend DNA but failed to proceed further in oxoG recognition. Modeling of the base eversion with energy decomposition suggested that the wedge destabilizes the intrahelical base primarily through buckling both surrounding base pairs. Replacement of oxoG with abasic (AP) site rescued the activity, and calculations suggested that wedge insertion is not required for AP site destabilization and eversion. Our results suggest that Fpg, and possibly other DNA glycosylases, convert part of the binding energy into active destabilization of their substrates, using the energy differences between normal and damaged bases for fast substrate discrimination.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Gilboa R., Zharkov D.O., Golan G., Fernandes A.S., Gerchman S.E., Matz E., Kycia J.H., Grollman A.P., Shoham G. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J. Biol. Chem. 2002;277:19811–19816. - PubMed
-
- Fromme J.C., Verdine G.L. Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nat. Struct. Biol. 2002;9:544–552. - PubMed
-
- Fromme J.C., Verdine G.L. DNA lesion recognition by the bacterial repair enzyme MutM. J. Biol. Chem. 2003;278:51543–51548. - PubMed
-
- Banerjee A., Santos W.L., Verdine G.L. Structure of a DNA glycosylase searching for lesions. Science. 2006;311:1153–1157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

