Semantically linking in silico cancer models
- PMID: 25520553
- PMCID: PMC4260769
- DOI: 10.4137/CIN.S13895
Semantically linking in silico cancer models
Abstract
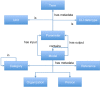
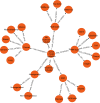
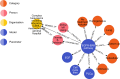
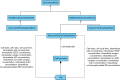
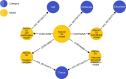
Multiscale models are commonplace in cancer modeling, where individual models acting on different biological scales are combined within a single, cohesive modeling framework. However, model composition gives rise to challenges in understanding interfaces and interactions between them. Based on specific domain expertise, typically these computational models are developed by separate research groups using different methodologies, programming languages, and parameters. This paper introduces a graph-based model for semantically linking computational cancer models via domain graphs that can help us better understand and explore combinations of models spanning multiple biological scales. We take the data model encoded by TumorML, an XML-based markup language for storing cancer models in online repositories, and transpose its model description elements into a graph-based representation. By taking such an approach, we can link domain models, such as controlled vocabularies, taxonomic schemes, and ontologies, with cancer model descriptions to better understand and explore relationships between models. The union of these graphs creates a connected property graph that links cancer models by categorizations, by computational compatibility, and by semantic interoperability, yielding a framework in which opportunities for exploration and discovery of combinations of models become possible.
Keywords: in silico oncology; model exploration; neo4j; property graphs; tumor modeling.
Figures
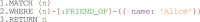


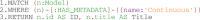

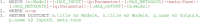
References
-
- Tracqui P. Biophysical models of tumour growth. Rep Prog Phys. 2009;72:56701.
-
- Walker DC, Southgate J. The virtual cell–a candidate co-ordinator for ‘middle-out’ modelling of biological systems. Brief Bioinform. 2009;10:450–61. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

