Localized, non-random differences in chromatin accessibility between homologous metaphase chromosomes
- PMID: 25520753
- PMCID: PMC4269072
- DOI: 10.1186/s13039-014-0070-y
Localized, non-random differences in chromatin accessibility between homologous metaphase chromosomes
Abstract
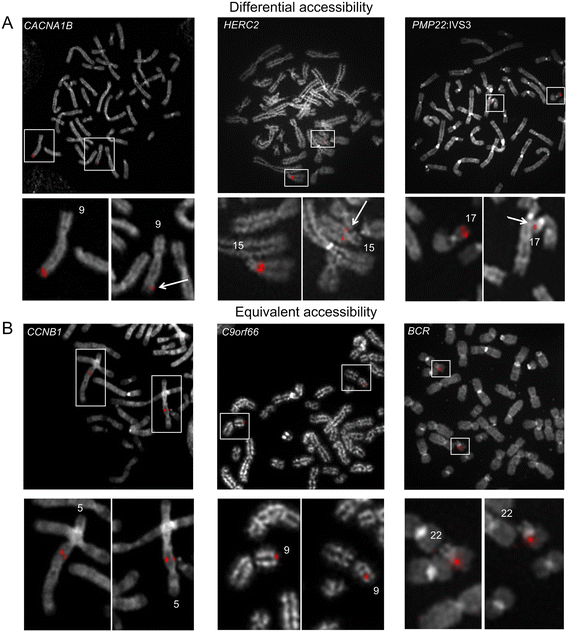
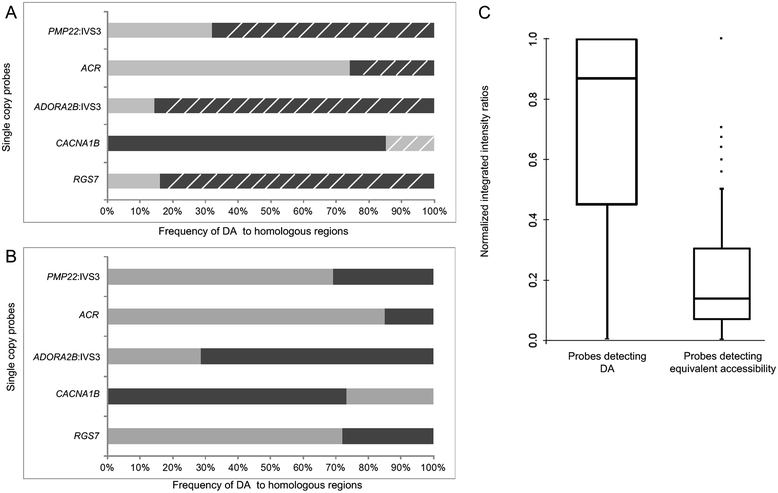
Background: Condensation differences along the lengths of homologous, mitotic metaphase chromosomes are well known. This study reports molecular cytogenetic data showing quantifiable localized differences in condensation between homologs that are related to differences in accessibility (DA) of associated DNA probe targets. Reproducible DA was observed for ~10% of locus-specific, short (1.5-5 kb) single copy DNA probes used in fluorescence in situ hybridization.
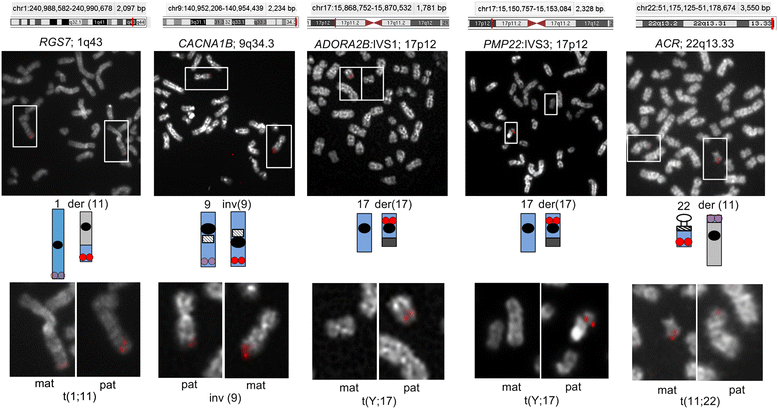
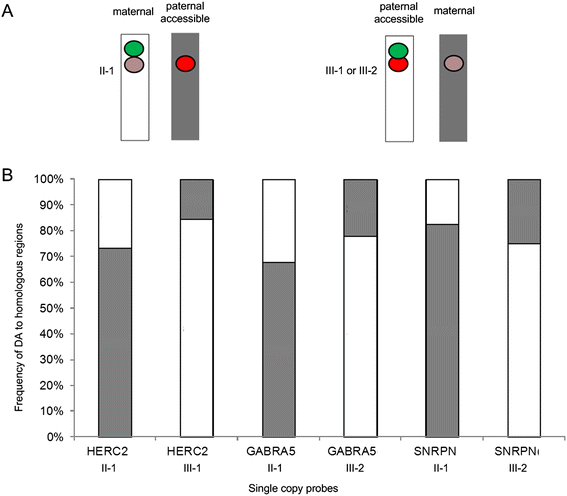
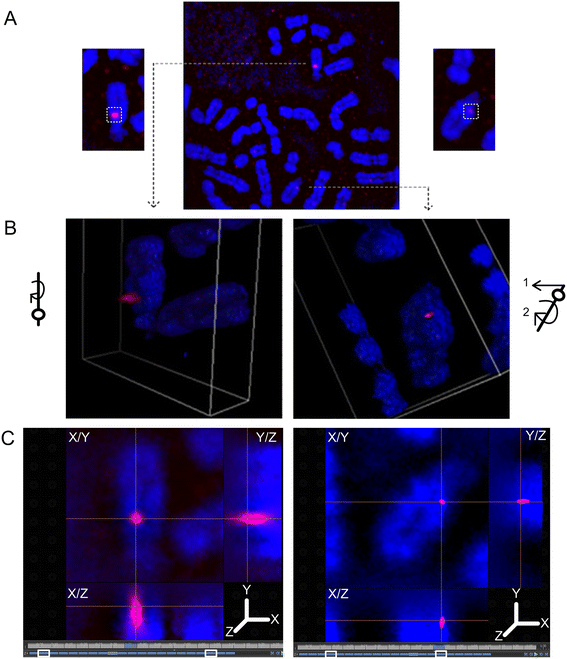
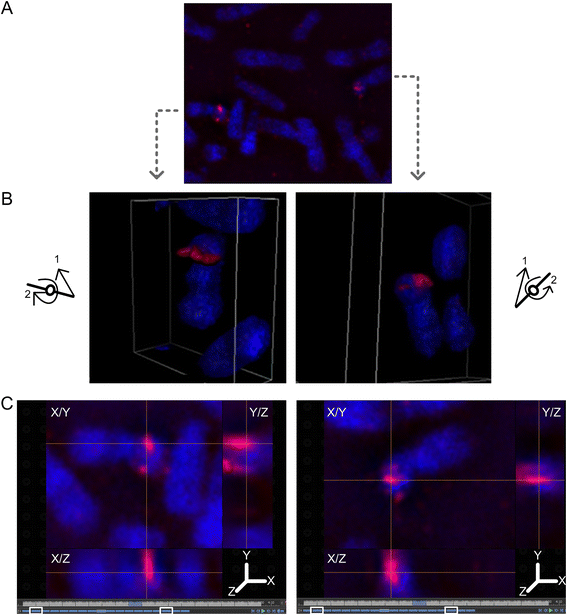
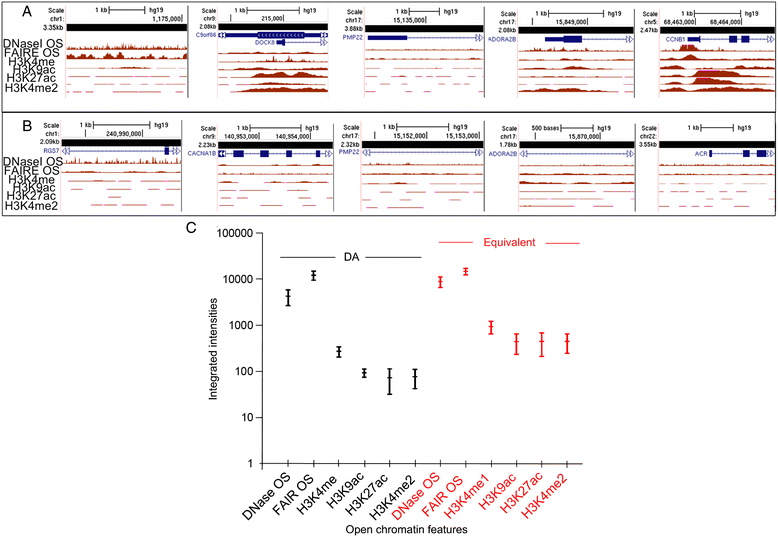
Results: Fourteen probes (from chromosomes 1, 5, 9, 11, 15, 17, 22) targeting genic and intergenic regions were developed and hybridized to cells from 10 individuals with cytogenetically-distinguishable homologs. Differences in hybridization between homologs were non-random for 8 genomic regions (RGS7, CACNA1B, GABRA5, SNRPN, HERC2, PMP22:IVS3, ADORA2B:IVS1, ACR) and were not unique to known imprinted domains or specific chromosomes. DNA probes within CCNB1, C9orf66, ADORA2B:Promoter-Ex1, PMP22:IVS4-Ex 5, and intergenic region 1p36.3 showed no DA (equivalent accessibility), while OPCML showed unbiased DA. To pinpoint probe locations, we performed 3D-structured illumination microscopy (3D-SIM). This showed that genomic regions with DA had 3.3-fold greater volumetric, integrated probe intensities and broad distributions of probe depths along axial and lateral axes of the 2 homologs, compared to a low copy probe target (NOMO1) with equivalent accessibility. Genomic regions with equivalent accessibility were also enriched for epigenetic marks of open interphase chromatin (DNase I HS, H3K27Ac, H3K4me1) to a greater extent than regions with DA.
Conclusions: This study provides evidence that DA is non-random and reproducible; it is locus specific, but not unique to known imprinted regions or specific chromosomes. Non-random DA was also shown to be heritable within a 2 generation family. DNA probe volume and depth measurements of hybridized metaphase chromosomes further show locus-specific chromatin accessibility differences by super-resolution 3D-SIM. Based on these data and the analysis of interphase epigenetic marks of genomic intervals with DA, we conclude that there are localized differences in compaction of homologs during mitotic metaphase and that these differences may arise during or preceding metaphase chromosome compaction. Our results suggest new directions for locus-specific structural analysis of metaphase chromosomes, motivated by the potential relationship of these findings to underlying epigenetic changes established during interphase.
Keywords: 3-D super resolution microscopy; Allelic differences; Differential chromatin accessibility; Homologous chromosome structure; Human mitotic chromosomes; Metaphase single copy FISH; Molecular cytogenetics.
Figures
Similar articles
-
Differentially accessible, single copy sequences form contiguous domains along metaphase chromosomes that are conserved among multiple tissues.Mol Cytogenet. 2021 Oct 20;14(1):49. doi: 10.1186/s13039-021-00567-w. Mol Cytogenet. 2021. PMID: 34670606 Free PMC article.
-
Reversing chromatin accessibility differences that distinguish homologous mitotic metaphase chromosomes.Mol Cytogenet. 2015 Aug 13;8:65. doi: 10.1186/s13039-015-0159-y. eCollection 2015. Mol Cytogenet. 2015. PMID: 26273322 Free PMC article.
-
Single-Molecule Micromanipulation and Super-Resolution Imaging Resolve Nanodomains Underlying Chromatin Folding in Mitotic Chromosomes.ACS Nano. 2022 May 24;16(5):8030-8039. doi: 10.1021/acsnano.2c01025. Epub 2022 Apr 29. ACS Nano. 2022. PMID: 35485433
-
Cytogenetic profiling using fluorescence in situ hybridization (FISH) and comparative genomic hybridization (CGH).J Cell Biochem Suppl. 1993;17G:139-43. doi: 10.1002/jcb.240531127. J Cell Biochem Suppl. 1993. PMID: 8007691 Review.
-
[Extending the capabilities of human chromosome analysis: from high-resolution banding to chromatin fiber-FISH].Hum Cell. 1997 Jun;10(2):121-34. Hum Cell. 1997. PMID: 9390269 Review. Japanese.
Cited by
-
Differentially accessible, single copy sequences form contiguous domains along metaphase chromosomes that are conserved among multiple tissues.Mol Cytogenet. 2021 Oct 20;14(1):49. doi: 10.1186/s13039-021-00567-w. Mol Cytogenet. 2021. PMID: 34670606 Free PMC article.
-
Immunolabelling of human metaphase chromosomes reveals the same banded distribution of histone H3 isoforms methylated at lysine 4 in primary lymphocytes and cultured cell lines.BMC Genet. 2015 Apr 29;16:44. doi: 10.1186/s12863-015-0200-5. BMC Genet. 2015. PMID: 25925961 Free PMC article.
-
Genome maintenance in the context of 4D chromatin condensation.Cell Mol Life Sci. 2016 Aug;73(16):3137-50. doi: 10.1007/s00018-016-2221-2. Epub 2016 Apr 20. Cell Mol Life Sci. 2016. PMID: 27098512 Free PMC article. Review.
-
Walking along chromosomes with super-resolution imaging, contact maps, and integrative modeling.PLoS Genet. 2018 Dec 26;14(12):e1007872. doi: 10.1371/journal.pgen.1007872. eCollection 2018 Dec. PLoS Genet. 2018. PMID: 30586358 Free PMC article.
-
High-resolution tyramide-FISH mapping of markers tightly linked to the male-fertility restoration (Ms) locus of onion.Theor Appl Genet. 2016 Mar;129(3):535-45. doi: 10.1007/s00122-015-2646-2. Epub 2015 Dec 24. Theor Appl Genet. 2016. PMID: 26704420
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

