Combining spatial and chemical information for clustering pharmacophores
- PMID: 25521061
- PMCID: PMC4290656
- DOI: 10.1186/1471-2105-15-S16-S5
Combining spatial and chemical information for clustering pharmacophores
Abstract
Background: A pharmacophore model consists of a group of chemical features arranged in three-dimensional space that can be used to represent the biological activities of the described molecules. Clustering of molecular interactions of ligands on the basis of their pharmacophore similarity provides an approach for investigating how diverse ligands can bind to a specific receptor site or different receptor sites with similar or dissimilar binding affinities. However, efficient clustering of pharmacophore models in three-dimensional space is currently a challenge.
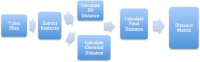
Results: We have developed a pharmacophore-assisted Iterative Closest Point (ICP) method that is able to group pharmacophores in a manner relevant to their biochemical properties, such as binding specificity etc. The implementation of the method takes pharmacophore files as input and produces distance matrices. The method integrates both alignment-dependent and alignment-independent concepts.
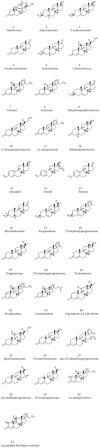
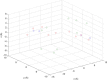
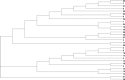
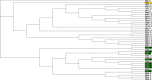
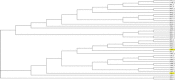
Conclusions: We apply our three-dimensional pharmacophore clustering method to two sets of experimental data, including 31 globulin-binding steroids and 4 groups of selected antibody-antigen complexes. Results are translated from distance matrices to Newick format and visualised using dendrograms. For the steroid dataset, the resulting classification of ligands shows good correspondence with existing classifications. For the antigen-antibody datasets, the classification of antigens reflects both antigen type and binding antibody. Overall the method runs quickly and accurately for classifying the data based on their binding affinities or antigens.
Figures




References
-
- Wermuth G, Ganellin CR, Lindberg P, Mitscher LA. Glossary of terms used in medicinal chemistry (IUPAC Recommendations 1998) Pure Appl Chem. 1998;70(5):1129–1143.
-
- Wolber G, Dornhofer AA, Langer T. Efficient overlay of small organic molecules using 3D pharmacophores. Journal of computer-aided molecular design. 2006;20(12):773–788. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

