Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death
- PMID: 25521377
- PMCID: PMC4632245
- DOI: 10.1016/j.neuron.2014.12.010
Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death
Abstract
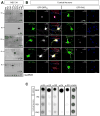
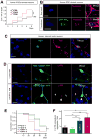
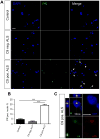
Expanded GGGGCC (G4C2) nucleotide repeats within the C9ORF72 gene are the most common genetic mutation associated with both amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Sense and antisense transcripts of these expansions are translated to form five dipeptide repeat proteins (DRPs). We employed primary cortical and motor neuron cultures, live-cell imaging, and transgenic fly models and found that the arginine-rich dipeptides, in particular Proline-Arginine (PR), are potently neurotoxic. Factors that anticipated their neurotoxicity included aggregation in nucleoli, decreased number of processing bodies, and stress granule formation, implying global translational dysregulation as path accountable for toxicity. Nuclear PR aggregates were also found in human induced motor neurons and postmortem spinal cord tissues from C9ORF72 ALS and ALS/FTD patients. Intronic G4C2 transcripts, but not loss of C9ORF72 protein, are also toxic to motor and cortical neurons. Interestingly, G4C2 transcript-mediated neurotoxicity synergizes with that of PR aggregates, suggesting convergence of mechanisms.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures





References
-
- Byrne S, Elamin M, Bede P, Shatunov A, Walsh C, Corr B, Heverin M, Jordan N, Kenna K, Lynch C, et al. Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: a population-based cohort study. Lancet Neurol. 2012;11:232–240. - PMC - PubMed
-
- Ciura S, Lattante S, Le Ber I, Latouche M, Tostivint H, Brice A, Kabashi E. Loss of function of C9orf72 causes motor deficits in a zebrafish model of Amyotrophic Lateral Sclerosis. Ann Neurol 2013 - PubMed
-
- Daigle JG, Lanson NA, Jr, Smith RB, Casci I, Maltare A, Monaghan J, Nichols CD, Kryndushkin D, Shewmaker F, Pandey UB. RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations. Hum Mol Genet. 2013;22:1193–1205. - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

