dCaP: detecting differential binding events in multiple conditions and proteins
- PMID: 25522020
- PMCID: PMC4290593
- DOI: 10.1186/1471-2164-15-S9-S12
dCaP: detecting differential binding events in multiple conditions and proteins
Abstract
Background: Current ChIP-seq studies are interested in comparing multiple epigenetic profiles across several cell types and tissues simultaneously for studying constitutive and differential regulation. Simultaneous analysis of multiple epigenetic features in many samples can gain substantial power and specificity than analyzing individual features and/or samples separately. Yet there are currently few tools can perform joint inference of constitutive and differential regulation in multi-feature-multi-condition contexts with statistical testing. Existing tools either test regulatory variation for one factor in multiple samples at a time, or for multiple factors in one or two samples. Many of them only identify binary rather than quantitative variation, which are sensitive to threshold choices.
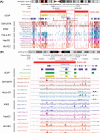
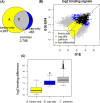
Results: We propose a novel and powerful method called dCaP for simultaneously detecting constitutive and differential regulation of multiple epigenetic factors in multiple samples. Using simulation, we demonstrate the superior power of dCaP compared to existing methods. We then apply dCaP to two datasets from human and mouse ENCODE projects to demonstrate its utility. We show in the human dataset that the cell-type specific regulatory loci detected by dCaP are significantly enriched near genes with cell-type specific functions and disease relevance. We further show in the mouse dataset that dCaP captures genomic regions showing significant signal variations for TAL1 occupancy between two mouse erythroid cell lines. The novel TAL1 occupancy loci detected only by dCaP are highly enriched with GATA1 occupancy and differential gene expression, while those detected only by other methods are not.
Conclusions: Here, we developed a novel approach to utilize the cooperative property of proteins to detect differential binding given multivariate ChIP-seq samples to provide better power, aiming for complementing existing approaches and providing new insights in the method development in this field.
Figures





Similar articles
-
A novel method for predicting cell abundance based on single-cell RNA-seq data.BMC Bioinformatics. 2021 Aug 25;22(Suppl 9):281. doi: 10.1186/s12859-021-04187-4. BMC Bioinformatics. 2021. PMID: 34433409 Free PMC article.
-
Dynamic shifts in occupancy by TAL1 are guided by GATA factors and drive large-scale reprogramming of gene expression during hematopoiesis.Genome Res. 2014 Dec;24(12):1945-62. doi: 10.1101/gr.164830.113. Epub 2014 Oct 15. Genome Res. 2014. PMID: 25319994 Free PMC article.
-
Condensin II subunit dCAP-D3 restricts retrotransposon mobilization in Drosophila somatic cells.PLoS Genet. 2013 Oct;9(10):e1003879. doi: 10.1371/journal.pgen.1003879. Epub 2013 Oct 31. PLoS Genet. 2013. PMID: 24204294 Free PMC article.
-
Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond.Cell Cycle. 2014;13(18):2847-52. doi: 10.4161/15384101.2014.949201. Cell Cycle. 2014. PMID: 25486472 Free PMC article. Review.
-
Epigenomics in stress tolerance of plants under the climate change.Mol Biol Rep. 2023 Jul;50(7):6201-6216. doi: 10.1007/s11033-023-08539-6. Epub 2023 Jun 9. Mol Biol Rep. 2023. PMID: 37294468 Review.
Cited by
-
InCoB2014: mining biological data from genomics for transforming industry and health.BMC Genomics. 2014;15 Suppl 9(Suppl 9):I1. doi: 10.1186/1471-2164-15-S9-I1. Epub 2014 Dec 8. BMC Genomics. 2014. PMID: 25521539 Free PMC article.
-
A MAD-Bayes Algorithm for State-Space Inference and Clustering with Application to Querying Large Collections of ChIP-Seq Data Sets.J Comput Biol. 2017 Jun;24(6):472-485. doi: 10.1089/cmb.2016.0138. Epub 2016 Nov 11. J Comput Biol. 2017. PMID: 27835030 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

