An integrative model of multi-organ drug-induced toxicity prediction using gene-expression data
- PMID: 25522097
- PMCID: PMC4290650
- DOI: 10.1186/1471-2105-15-S16-S2
An integrative model of multi-organ drug-induced toxicity prediction using gene-expression data
Abstract
Background: In practice, some drugs produce a number of negative biological effects that can mitigate their effectiveness as a remedy. To address this issue, several studies have been performed for the prediction of drug-induced toxicity from gene-expression data, and a significant amount of work has been done on predicting limited drug-induced symptoms or single-organ toxicity. Since drugs often lead to some injuries in several organs like liver or kidney, however, it would be very useful to forecast the drug-induced injuries for multiple organs. Therefore, in this work, our aim was to develop a multi-organ toxicity prediction model using an integrative model of gene-expression data.
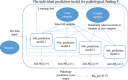
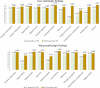
Results: To train our integrative model, we used 3708 in-vivo samples of gene-expression profiles exposed to one of 41 drugs related to 21 distinct physiological changes divided between liver and kidney (liver 11, kidney 10). Specifically, we used the gene-expression profiles to learn an ensemble classifier for each of 21 pathology prediction models. Subsequently, these classifiers were combined with weights to generate an integrative model for each pathological finding. The integrative model outputs the likeliness of presenting the trained pathology in a given test sample of gene-expression profile, called an integrative prediction score (IPS). For the evaluation of an integrative model, we estimated the prediction performance with the k-fold cross-validation. Our results demonstrate that the proposed integrative model is superior to individual pathology prediction models in predicting multi-organ drug-induced toxicities over all the targeted pathological findings. On average, the AUC of the integrative models was 88% while the AUC of individual pathology prediction models was 68%.
Conclusions: Not only does this integrative model produce comparable prediction performance to existing approaches, but also it produces very stable performance overall. In addition, our approach is easily expandable to a variety of other multi-organ toxicology applications.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

