Plasmid flux in Escherichia coli ST131 sublineages, analyzed by plasmid constellation network (PLACNET), a new method for plasmid reconstruction from whole genome sequences
- PMID: 25522143
- PMCID: PMC4270462
- DOI: 10.1371/journal.pgen.1004766
Plasmid flux in Escherichia coli ST131 sublineages, analyzed by plasmid constellation network (PLACNET), a new method for plasmid reconstruction from whole genome sequences
Abstract
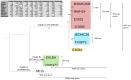
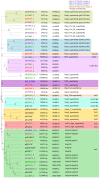
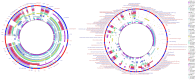
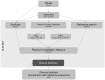
Bacterial whole genome sequence (WGS) methods are rapidly overtaking classical sequence analysis. Many bacterial sequencing projects focus on mobilome changes, since macroevolutionary events, such as the acquisition or loss of mobile genetic elements, mainly plasmids, play essential roles in adaptive evolution. Existing WGS analysis protocols do not assort contigs between plasmids and the main chromosome, thus hampering full analysis of plasmid sequences. We developed a method (called plasmid constellation networks or PLACNET) that identifies, visualizes and analyzes plasmids in WGS projects by creating a network of contig interactions, thus allowing comprehensive plasmid analysis within WGS datasets. The workflow of the method is based on three types of data: assembly information (including scaffold links and coverage), comparison to reference sequences and plasmid-diagnostic sequence features. The resulting network is pruned by expert analysis, to eliminate confounding data, and implemented in a Cytoscape-based graphic representation. To demonstrate PLACNET sensitivity and efficacy, the plasmidome of the Escherichia coli lineage ST131 was analyzed. ST131 is a globally spread clonal group of extraintestinal pathogenic E. coli (ExPEC), comprising different sublineages with ability to acquire and spread antibiotic resistance and virulence genes via plasmids. Results show that plasmids flux in the evolution of this lineage, which is wide open for plasmid exchange. MOBF12/IncF plasmids were pervasive, adding just by themselves more than 350 protein families to the ST131 pangenome. Nearly 50% of the most frequent γ-proteobacterial plasmid groups were found to be present in our limited sample of ten analyzed ST131 genomes, which represent the main ST131 sublineages.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

