Complete chloroplast genome of Macadamia integrifolia confirms the position of the Gondwanan early-diverging eudicot family Proteaceae
- PMID: 25522147
- PMCID: PMC4290595
- DOI: 10.1186/1471-2164-15-S9-S13
Complete chloroplast genome of Macadamia integrifolia confirms the position of the Gondwanan early-diverging eudicot family Proteaceae
Abstract
Background: Sequence data from the chloroplast genome have played a central role in elucidating the evolutionary history of flowering plants, Angiospermae. In the past decade, the number of complete chloroplast genomes has burgeoned, leading to well-supported angiosperm phylogenies. However, some relationships, particulary among early-diverging lineages, remain unresolved. The diverse Southern Hemisphere plant family Proteaceae arose on the ancient supercontinent Gondwana early in angiosperm history and is a model group for adaptive radiation in response to changing climatic conditions. Genomic resources for the family are limited, and until now it is one of the few early-diverging 'basal eudicot' lineages not represented in chloroplast phylogenomic analyses.
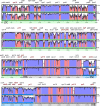
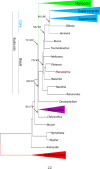
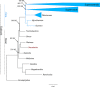
Results: The chloroplast genome of the Australian nut crop tree Macadamia integrifolia was assembled de novo from Illumina paired-end sequence reads. Three contigs, corresponding to a collapsed inverted repeat, a large and a small single copy region were identified, and used for genome reconstruction. The complete genome is 159,714 bp in length and was assembled at deep coverage (3.29 million reads; ~2000 x). Phylogenetic analyses based on 83-gene and inverted repeat region alignments, the largest sequence-rich datasets to include the basal eudicot family Proteaceae, provide strong support for a Proteales clade that includes Macadamia, Platanus and Nelumbo. Genome structure and content followed the ancestral angiosperm pattern and were highly conserved in the Proteales, whilst size differences were largely explained by the relative contraction of the single copy regions and expansion of the inverted repeats in Macadamia.
Conclusions: The Macadamia chloroplast genome presented here is the first in the Proteaceae, and confirms the placement of this family with the morphologically divergent Plantanaceae (plane tree family) and Nelumbonaceae (sacred lotus family) in the basal eudicot order Proteales. It provides a high-quality reference genome for future evolutionary studies and will be of benefit for taxon-rich phylogenomic analyses aimed at resolving relationships among early-diverging angiosperms, and more broadly across the plant tree of life.
Figures

References
-
- Palmer JD, Stein DB. Conservation of chloroplast genome structure among vascular plants. Curr Genet. 1986;10:823–833. doi: 10.1007/BF00418529. - DOI
-
- Jansen RK, Cai Z, Raubeson LA, Daniell H, dePamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW, Lee S, Peery R, McNeal JR, Kuehl JV, Boore JL. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. - DOI - PMC - PubMed
-
- Moore MJ, Hassan N, Gitzendanner MA, Bruenn RA, Croley M, Vandeventer A, Horn JW, Dhingra A, Brockington SF, Latvis M, Ramdial J, Alexandre R, Peidrahita A, Xi Z, Davis CC, Soltis PS, Soltis DE. Phylogenetic analysis of the plastid inverted repeat for 244 species: insights into deeper-level angiosperm relationships from a long, slowly evolving sequence region. Int J Plant Sci. 2011;172(4):541–558. doi: 10.1086/658923. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

