RNA recognition and stress granule formation by TIA proteins
- PMID: 25522169
- PMCID: PMC4284772
- DOI: 10.3390/ijms151223377
RNA recognition and stress granule formation by TIA proteins
Abstract
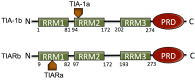
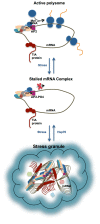
Stress granule (SG) formation is a primary mechanism through which gene expression is rapidly modulated when the eukaryotic cell undergoes cellular stresses (including heat, oxidative, viral infection, starvation). In particular, the sequestration of specifically targeted translationally stalled mRNAs into SGs limits the expression of a subset of genes, but allows the expression of heatshock proteins that have a protective effect in the cell. The importance of SGs is seen in several disease states in which SG function is disrupted. Fundamental to SG formation are the T cell restricted intracellular antigen (TIA) proteins (TIA-1 and TIA-1 related protein (TIAR)), that both directly bind to target RNA and self-associate to seed the formation of SGs. Here a summary is provided of the current understanding of the way in which TIA proteins target specific mRNA, and how TIA self-association is triggered under conditions of cellular stress.
Figures
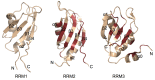
References
-
- Anderson P., Kedersha N. Stress granules. Curr. Biol. 2009;19:R397–R398. - PubMed
-
- Scheu S., Stetson D.B., Reinhardt R.L., Leber J.H., Mohrs M., Locksley R.M. Activation of the integrated stress response during T helper cell differentiation. Nat. Immunol. 2006;7:644–651. - PubMed
-
- Arimoto K., Fukuda H., Imajoh-Ohmi S., Saito H., Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 2008;10:1324–1332. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

