Molecular evolution of broadly neutralizing Llama antibodies to the CD4-binding site of HIV-1
- PMID: 25522326
- PMCID: PMC4270772
- DOI: 10.1371/journal.ppat.1004552
Molecular evolution of broadly neutralizing Llama antibodies to the CD4-binding site of HIV-1
Abstract
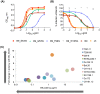
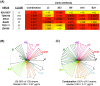
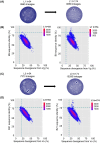
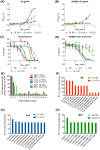
To date, no immunization of humans or animals has elicited broadly neutralizing sera able to prevent HIV-1 transmission; however, elicitation of broad and potent heavy chain only antibodies (HCAb) has previously been reported in llamas. In this study, the anti-HIV immune responses in immunized llamas were studied via deep sequencing analysis using broadly neutralizing monoclonal HCAbs as a guides. Distinct neutralizing antibody lineages were identified in each animal, including two defined by novel antibodies (as variable regions called VHH) identified by robotic screening of over 6000 clones. The combined application of five VHH against viruses from clades A, B, C and CRF_AG resulted in neutralization as potent as any of the VHH individually and a predicted 100% coverage with a median IC50 of 0.17 µg/ml for the panel of 60 viruses tested. Molecular analysis of the VHH repertoires of two sets of immunized animals showed that each neutralizing lineage was only observed following immunization, demonstrating that they were elicited de novo. Our results show that immunization can induce potent and broadly neutralizing antibodies in llamas with features similar to human antibodies and provide a framework to analyze the effectiveness of immunization protocols.
Conflict of interest statement
LR and TV are employed by a commercial company, QVQ B.V. This does not alter our adherence to all PLOS Pathogens policies on sharing data and materials.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

