Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature
- PMID: 25524025
- PMCID: PMC4284803
- DOI: 10.1038/ncomms6812
Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature
Abstract
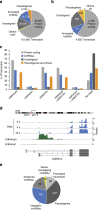
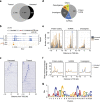
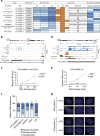
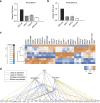
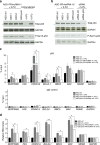
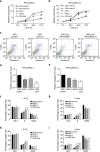
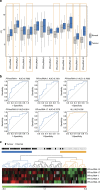
Despite the inarguable relevance of p53 in cancer, genome-wide studies relating endogenous p53 activity to the expression of lncRNAs in human cells are still missing. Here, by integrating RNA-seq with p53 ChIP-seq analyses of a human cancer cell line under DNA damage, we define a high-confidence set of 18 lncRNAs that are p53 transcriptional targets. We demonstrate that two of the p53-regulated lncRNAs are required for the efficient binding of p53 to some of its target genes, modulating the p53 transcriptional network and contributing to apoptosis induction by DNA damage. We also show that the expression of p53-lncRNAs is lowered in colorectal cancer samples, constituting a tumour suppressor signature with high diagnostic power. Thus, p53-regulated lncRNAs establish a positive regulatory feedback loop that enhances p53 tumour suppressor activity. Furthermore, the signature defined by p53-regulated lncRNAs supports their potential use in the clinic as biomarkers and therapeutic targets.
Figures
References
-
- Menendez D., Inga A. & Resnick M. A. The expanding universe of p53 targets. Nat. Rev. Cancer 9, 724–737 (2009). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

