Algebraic Statistical Model for Biochemical Network Dynamics Inference
- PMID: 25525612
- PMCID: PMC4267476
- DOI: 10.1166/jcsmd.2013.1032
Algebraic Statistical Model for Biochemical Network Dynamics Inference
Abstract
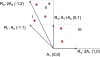
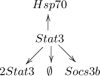
With modern molecular quantification methods, like, for instance, high throughput sequencing, biologists may perform multiple complex experiments and collect longitudinal data on RNA and DNA concentrations. Such data may be then used to infer cellular level interactions between the molecular entities of interest. One method which formalizes such inference is the stoichiometric algebraic statistical model (SASM) of [2] which allows to analyze the so-called conic (or single source) networks. Despite its intuitive appeal, up until now the SASM has been only heuristically studied on few simple examples. The current paper provides a more formal mathematical treatment of the SASM, expanding the original model to a wider class of reaction systems decomposable into multiple conic subnetworks. In particular, it is proved here that on such networks the SASM enjoys the so-called sparsistency property, that is, it asymptotically (with the number of observed network trajectories) discards the false interactions by setting their reaction rates to zero. For illustration, we apply the extended SASM to in silico data from a generic decomposable network as well as to biological data from an experimental search for a possible transcription factor for the heat shock protein 70 (Hsp70) in the zebrafish retina.
Keywords: Algebraic Statistical Model; Biochemical Networks; DNA- and RNA-based Technologies; Law of Mass Action; Parameter Inference; Systems Biology.
Conflict of interest statement
Figures

Similar articles
-
Statistical Model for Biochemical Network Inference.Commun Stat Simul Comput. 2013 Jan;42(1):121-137. doi: 10.1080/03610918.2011.633200. Epub 2012 Sep 26. Commun Stat Simul Comput. 2013. PMID: 23125476 Free PMC article.
-
An algebra-based method for inferring gene regulatory networks.BMC Syst Biol. 2014 Mar 26;8:37. doi: 10.1186/1752-0509-8-37. BMC Syst Biol. 2014. PMID: 24669835 Free PMC article.
-
Biological Network Inference and analysis using SEBINI and CABIN.Methods Mol Biol. 2009;541:551-76. doi: 10.1007/978-1-59745-243-4_24. Methods Mol Biol. 2009. PMID: 19381531 Review.
-
Inferring biochemical reaction pathways: the case of the gemcitabine pharmacokinetics.BMC Syst Biol. 2012 May 28;6:51. doi: 10.1186/1752-0509-6-51. BMC Syst Biol. 2012. PMID: 22640931 Free PMC article.
-
On protocols and measures for the validation of supervised methods for the inference of biological networks.Front Genet. 2013 Dec 3;4:262. doi: 10.3389/fgene.2013.00262. Front Genet. 2013. PMID: 24348517 Free PMC article. Review.
Cited by
-
Reverse engineering gene networks using global-local shrinkage rules.Interface Focus. 2020 Feb 6;10(1):20190049. doi: 10.1098/rsfs.2019.0049. Epub 2019 Dec 13. Interface Focus. 2020. PMID: 31897291 Free PMC article.
References
-
- Close Jennie L, Gumuscu Burak, Reh Thomas A. Retinal neurons regulate proliferation of postnatal progenitors and Müller glia in the rat retina via TGFβ signaling. Development. 2005;132(13):3015–3026. - PubMed
-
- Gillespie Daniel T. A rigorous derivation of the chemical master equation. Physica A: Statistical Mechanics and its Applications. 1992;188(1):404–425.
-
- Kim Jaejik, Rempala Grzegorz A. Bioreactor simulation software. 2013 URL https://neyman.cph.ohio-state.edu/bioreactor.html.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
