Architectural insight into inovirus-associated vectors (IAVs) and development of IAV-based vaccines inducing humoral and cellular responses: implications in HIV-1 vaccines
- PMID: 25525909
- PMCID: PMC4276942
- DOI: 10.3390/v6125047
Architectural insight into inovirus-associated vectors (IAVs) and development of IAV-based vaccines inducing humoral and cellular responses: implications in HIV-1 vaccines
Abstract
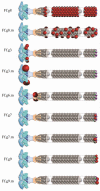
Inovirus-associated vectors (IAVs) are engineered, non-lytic, filamentous bacteriophages that are assembled primarily from thousands of copies of the major coat protein gp8 and just five copies of each of the four minor coat proteins gp3, gp6, gp7 and gp9. Inovirus display studies have shown that the architecture of inoviruses makes all coat proteins of the inoviral particle accessible to the outside. This particular feature of IAVs allows foreign antigenic peptides to be displayed on the outer surface of the virion fused to its coat proteins and for more than two decades has been exploited in many applications including antibody or peptide display libraries, drug design, and vaccine development against infectious and non-infectious diseases. As vaccine carriers, IAVs have been shown to elicit both a cellular and humoral response against various pathogens through the display of antibody epitopes on their coat proteins. Despite their high immunogenicity, the goal of developing an effective vaccine against HIV-1 has not yet materialized. One possible limitation of previous efforts was the use of broadly neutralizing antibodies, which exhibited autoreactivity properties. In the past five years, however, new, more potent broadly neutralizing antibodies that do not exhibit autoreactivity properties have been isolated from HIV-1 infected individuals, suggesting that vaccination strategies aimed at producing such broadly neutralizing antibodies may confer protection against infection. The utilization of these new, broadly neutralizing antibodies in combination with the architectural traits of IAVs have driven the current developments in the design of an inovirus-based vaccine against HIV-1. This article reviews the applications of IAVs in vaccine development, with particular emphasis on the design of inoviral-based vaccines against HIV-1.
Figures


References
-
- Day L.A. Inoviruses. In: Mahy B.W.J., Regenmortel M.H.V.V., editors. Encyclopedia of Virology. 3rd ed. Academic Press; Oxford, UK: 2008. pp. 117–124.
-
- Marvin D.A., Symmons M.F., Straus S.K. Structure and assembly of filamentous bacteriophages. Prog. Biophys. Mol. Biol. 2014;114:80–122. - PubMed
-
- Day L.A., Marzec C.J., Reisberg S.A., Casadevall A. DNA packing in filamentous bacteriophages. Annu. Rev. Biophys. Biophys. Chem. 1988;17:509–539. - PubMed
-
- Rakonjac J., Bennett N.J., Spagnuolo J., Gagic D., Russel M. Filamentous bacteriophage: Biology, phage display and nanotechnology applications. Curr. Issues Mol. Biol. 2011;13:51–76. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

