The role of cytochromes p450 and aldo-keto reductases in prognosis of breast carcinoma patients
- PMID: 25526449
- PMCID: PMC4603110
- DOI: 10.1097/MD.0000000000000255
The role of cytochromes p450 and aldo-keto reductases in prognosis of breast carcinoma patients
Abstract
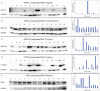
Metabolism of anticancer drugs affects their antitumor effects. This study has investigated the associations of gene expression of enzymes metabolizing anticancer drugs with therapy response and survival of breast carcinoma patients. Gene expression of 13 aldo-keto reductases (AKRs), carbonyl reductase 1, and 10 cytochromes P450 (CYPs) was assessed using quantitative real-time polymerase chain reaction in tumors and paired adjacent nonneoplastic tissues from 68 posttreatment breast carcinoma patients. Eleven candidate genes were then evaluated in an independent series of 50 pretreatment patients. Protein expression of the most significant genes was confirmed by immunoblotting. AKR1A1 was significantly overexpressed and AKR1C1-4, KCNAB1, CYP2C19, CYP3A4, and CYP3A5 downregulated in tumors compared with control nonneoplastic tissues after correction for multiple testing. Significant association of CYP2B6 transcript levels in tumors with expression of hormonal receptors was found in the posttreatment set and replicated in the pretreatment set of patients. Significantly higher intratumoral levels of AKR1C1, AKR1C2, or CYP2W1 were found in responders to neoadjuvant chemotherapy compared with nonresponders. Patients with high AKR7A3 or CYP2B6 levels in the pretreatment set had significantly longer disease-free survival than patients with low levels. Protein products of AKR1C1, AKR1C2, AKR7A3, CYP3A4, and carbonyl reductase (CBR1) were found in tumors and those of AKR1C1, AKR7A3, and CBR1 correlated with their transcript levels. Small interfering RNA-directed knockdown of AKR1C2 or vector-mediated upregulation of CYP3A4 in MDA-MB-231 model cell line had no effect on cell proliferation after paclitaxel treatment in vitro. Prognostic and predictive roles of drug-metabolizing enzymes strikingly differ between posttreatment and pretreatment breast carcinoma patients. Mechanisms of action of AKR1C2, AKR7A3, CYP2B6, CYP3A4, and CBR1 should continue to be further followed in breast carcinoma patients and models.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures





References
-
- Ferlay J, Shin HR, Bray F, et al. GLOBOCAN 2008, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 10. Lyon, France: International Agency for Research on Cancer; 2010.
-
- Baguley BC. Multiple drug resistance mechanisms in cancer. Mol Biotechnol 2010; 46:308–316. - PubMed
-
- Johansson I, Ingelman-Sundberg M. Genetic polymorphism and toxicology with emphasis on cytochrome P450. Toxicol Sci 2011; 120:1–13. - PubMed
-
- Roy P, Yu LJ, Crespi CL, et al. Development of a substrate-activity based approach to identify the major human liver P-450 catalysts of cyclophosphamide and ifosfamide activation based on cDNA-expressed activities and liver microsomal P-450 profiles. Drug Metab Dispos 1999; 27:655–666. - PubMed
-
- Chang TK, Weber GF, Crespi CL, et al. Differential activation of cyclophosphamide and ifosphamide by cytochromes P-450 2B and 3A in human liver microsomes. Cancer Res 1993; 53:5629–5637. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

