What can crystal structures of aminergic receptors tell us about designing subtype-selective ligands?
- PMID: 25527701
- PMCID: PMC4279073
- DOI: 10.1124/pr.114.009944
What can crystal structures of aminergic receptors tell us about designing subtype-selective ligands?
Abstract
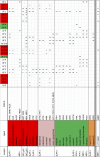
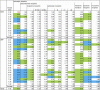
G protein-coupled receptors (GPCRs) are integral membrane proteins that represent an important class of drug targets. In particular, aminergic GPCRs interact with a significant portion of drugs currently on the market. However, most drugs that target these receptors are associated with undesirable side effects, which are due in part to promiscuous interactions with close homologs of the intended target receptors. Here, based on a systematic analysis of all 37 of the currently available high-resolution crystal structures of aminergic GPCRs, we review structural elements that contribute to and can be exploited for designing subtype-selective compounds. We describe the roles of secondary binding pockets (SBPs), as well as differences in ligand entry pathways to the orthosteric binding site, in determining selectivity. In addition, using the available crystal structures, we have identified conformational changes in the SBPs that are associated with receptor activation and explore the implications of these changes for the rational development of selective ligands with tailored efficacy.
U.S. Government work not protected by U.S. copyright.
Figures


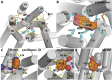
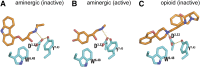


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

