The root hair "infectome" of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for Auxin signaling in rhizobial infection
- PMID: 25527707
- PMCID: PMC4311213
- DOI: 10.1105/tpc.114.133496
The root hair "infectome" of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for Auxin signaling in rhizobial infection
Abstract
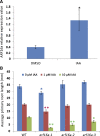
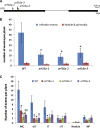
Nitrogen-fixing rhizobia colonize legume roots via plant-made intracellular infection threads. Genetics has identified some genes involved but has not provided sufficient detail to understand requirements for infection thread development. Therefore, we transcriptionally profiled Medicago truncatula root hairs prior to and during the initial stages of infection. This revealed changes in the responses to plant hormones, most notably auxin, strigolactone, gibberellic acid, and brassinosteroids. Several auxin responsive genes, including the ortholog of Arabidopsis thaliana Auxin Response Factor 16, were induced at infection sites and in nodule primordia, and mutation of ARF16a reduced rhizobial infection. Associated with the induction of auxin signaling genes, there was increased expression of cell cycle genes including an A-type cyclin and a subunit of the anaphase promoting complex. There was also induction of several chalcone O-methyltransferases involved in the synthesis of an inducer of Sinorhizobium meliloti nod genes, as well as a gene associated with Nod factor degradation, suggesting both positive and negative feedback loops that control Nod factor levels during rhizobial infection. We conclude that the onset of infection is associated with reactivation of the cell cycle as well as increased expression of genes required for hormone and flavonoid biosynthesis and that the regulation of auxin signaling is necessary for initiation of rhizobial infection threads.
© 2014 American Society of Plant Biologists. All rights reserved.
Figures




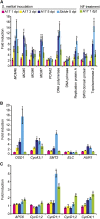
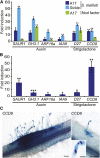

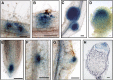
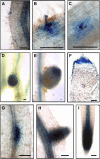


Comment in
-
Rhizobial root hair infection requires auxin signaling.Trends Plant Sci. 2015 Jun;20(6):332-4. doi: 10.1016/j.tplants.2015.04.004. Epub 2015 Apr 23. Trends Plant Sci. 2015. PMID: 25920666
References
-
- Akiyama K., Matsuzaki K., Hayashi H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827. - PubMed
-
- Allen N.S., Bennett M.N., Cox D.N., Shipley A., Ehrhardt D.W., Long S.R. (1994). Effects of Nod factors on alfalfa root hair Ca++ and H+ currents and on cytoskeletal behaviour. In Advances in Molecular Genetics of Plant-Microbe Interactions, Vol. 3. M.J. Daniels, J.A. Downie, and A.E. Osbourn, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 107–113.
-
- Anisimova M., Gascuel O. (2006). Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 55: 539–552. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/J/00000603/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/J/000CA282/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/G023832/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/L010305/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/J/00000012/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

