The liverwort Pellia endiviifolia shares microtranscriptomic traits that are common to green algae and land plants
- PMID: 25530158
- PMCID: PMC4368373
- DOI: 10.1111/nph.13220
The liverwort Pellia endiviifolia shares microtranscriptomic traits that are common to green algae and land plants
Abstract
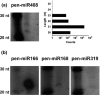
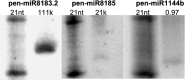
Liverworts are the most basal group of extant land plants. Nonetheless, the molecular biology of liverworts is poorly understood. Gene expression has been studied in only one species, Marchantia polymorpha. In particular, no microRNA (miRNA) sequences from liverworts have been reported. Here, Illumina-based next-generation sequencing was employed to identify small RNAs, and analyze the transcriptome and the degradome of Pellia endiviifolia. Three hundred and eleven conserved miRNA plant families were identified, and 42 new liverwort-specific miRNAs were discovered. The RNA degradome analysis revealed that target mRNAs of only three miRNAs (miR160, miR166, and miR408) have been conserved between liverworts and other land plants. New targets were identified for the remaining conserved miRNAs. Moreover, the analysis of the degradome permitted the identification of targets for 13 novel liverwort-specific miRNAs. Interestingly, three of the liverwort microRNAs show high similarity to previously reported miRNAs from Chlamydomonas reinhardtii. This is the first observation of miRNAs that exist both in a representative alga and in the liverwort P. endiviifolia but are not present in land plants. The results of the analysis of the P. endivifolia microtranscriptome support the conclusions of previous studies that placed liverworts at the root of the land plant evolutionary tree of life.
Keywords: Pellia endiviifolia; basal lineage; land plants; liverwort; miRNA precursors; miRNA targets; microRNA (miRNA) genes; microtranscriptome.
© 2014 The Authors. New Phytologist © 2014 New Phytologist Trust.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- SRA/SRP043263
- SRA/SRP048691
LinkOut - more resources
Full Text Sources
Other Literature Sources

