Genotype shift in human coronavirus OC43 and emergence of a novel genotype by natural recombination
- PMID: 25530469
- PMCID: PMC7112537
- DOI: 10.1016/j.jinf.2014.12.005
Genotype shift in human coronavirus OC43 and emergence of a novel genotype by natural recombination
Abstract
Background: Human coronavirus (HCoV) OC43 is the most prevalent HCoV in respiratory tract infections. Its molecular epidemiological characterization, particularly the genotyping, was poorly addressed.
Methods: The full-length spike (S), RNA-dependent RNA polymerase (RdRp), and nucleocapsid (N) genes were amplified from each respiratory sample collected from 65 HCoV-OC43-positive patients between 2005 and 2012. Genotypes were determined by phylogenetic analysis. Recombination was analyzed based on full-length viral genome sequences. Clinical manifestations of each HCoV genotype infection were compared by reviewing clinical records.
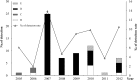
Results: Sixty of these 65 samples belong to genotypes B, C and D. The remaining five strains had incongruent positions in the phylogenetic trees of the S, RdRp and N genes, suggesting a novel genotype emerging, designated as genotype E. Whole genome sequencing and bootscan analysis indicated that genotype E is generated by recombination between genotypes B, C and D. Temporal analysis revealed a sequential genotype replacement of C, B, D and E over the study period with genotype D being the dominant genotype since 2007. The novel genotype E was only detected in children younger than three years suffering from lower respiratory tract infections.
Conclusions: Our results suggest that HCoV-OC43 genotypes are evolving. Such genotype shift may be an adapting mechanism for HCoV-OC43 maintaining its epidemic.
Keywords: Genotype; Human coronavirus OC43; Molecular epidemiology; Recombination; Respiratory infection.
Copyright © 2014 The British Infection Association. Published by Elsevier Ltd. All rights reserved.
Figures




References
-
- Masters P.S., Perlman S. Coronaviridae. In: Knipe D.M., Howley P.M., editors. Fields virology. 6th ed. Lippincott Williams &Wilkins; Philadelphia: 2013. pp. 825–854.
-
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86(7):3995–4008. - PMC - PubMed
-
- International Committee on Taxonomy of Viruses. Virus taxonomy: 2013 Release. http://ictvonline.org/virusTaxonomy.asp [accessed 20.05.14].
-
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814–1820. - PubMed
-
- Bialek S.R., Allen D., Alvarado-Ramy F., Arthur R., Balajee A., Bell D. First confirmed cases of middle east respiratory syndrome coronavirus (MERS-CoV) infection in the United States, updated information on the epidemiology of MERS-CoV infection, and guidance for the public, clinicians, and public health authorities – May 2014. MMWR Morb Mortal Wkly Rep. 2014;63(19):431–436. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

