Sequence conservation, phylogenetic relationships, and expression profiles of nondigestive serine proteases and serine protease homologs in Manduca sexta
- PMID: 25530503
- PMCID: PMC4474797
- DOI: 10.1016/j.ibmb.2014.10.006
Sequence conservation, phylogenetic relationships, and expression profiles of nondigestive serine proteases and serine protease homologs in Manduca sexta
Abstract
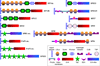
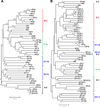
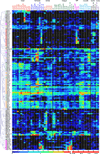
Serine protease (SP) and serine protease homolog (SPH) genes in insects encode a large family of proteins involved in digestion, development, immunity, and other processes. While 68 digestive SPs and their close homologs are reported in a companion paper (Kuwar et al., in preparation), we have identified 125 other SPs/SPHs in Manduca sexta and studied their structure, evolution, and expression. Fifty-two of them contain cystine-stabilized structures for molecular recognition, including clip, LDLa, Sushi, Wonton, TSP, CUB, Frizzle, and SR domains. There are nineteen groups of genes evolved from relatively recent gene duplication and sequence divergence. Thirty-five SPs and seven SPHs contain 1, 2 or 5 clip domains. Multiple sequence alignment and molecular modeling of the 54 clip domains have revealed structural diversity of these regulatory modules. Sequence comparison with their homologs in Drosophila melanogaster, Anopheles gambiae and Tribolium castaneum allows us to classify them into five subfamilies: A are SPHs with 1 or 5 group-3 clip domains, B are SPs with 1 or 2 group-2 clip domains, C, D1 and D2 are SPs with a single clip domain in group-1a, 1b and 1c, respectively. We have classified into six categories the 125 expression profiles of SP-related proteins in fat body, brain, midgut, Malpighian tubule, testis, and ovary at different stages, suggesting that they participate in various physiological processes. Through RNA-Seq-based gene annotation and expression profiling, as well as intragenomic sequence comparisons, we have established a framework of information for future biochemical research of nondigestive SPs and SPHs in this model species.
Keywords: Clip domain; Hemolymph protein; Insect immunity; Phylogenetic analysis; RNA-Seq.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures


Similar articles
-
Serine protease-related proteins in the malaria mosquito, Anopheles gambiae.Insect Biochem Mol Biol. 2017 Sep;88:48-62. doi: 10.1016/j.ibmb.2017.07.008. Epub 2017 Aug 2. Insect Biochem Mol Biol. 2017. PMID: 28780069 Free PMC article.
-
Comparative analysis of serine protease-related genes in the honey bee genome: possible involvement in embryonic development and innate immunity.Insect Mol Biol. 2006 Oct;15(5):603-14. doi: 10.1111/j.1365-2583.2006.00684.x. Insect Mol Biol. 2006. PMID: 17069636 Free PMC article.
-
Genome-wide identification and expression profiling of serine proteases and homologs in the diamondback moth, Plutella xylostella (L.).BMC Genomics. 2015 Dec 10;16:1054. doi: 10.1186/s12864-015-2243-4. BMC Genomics. 2015. PMID: 26653876 Free PMC article.
-
Drosophila melanogaster clip-domain serine proteases: Structure, function and regulation.Biochimie. 2016 Mar;122:255-69. doi: 10.1016/j.biochi.2015.10.007. Epub 2015 Oct 8. Biochimie. 2016. PMID: 26453810 Review.
-
Serine pseudoproteases in physiology and disease.FEBS J. 2023 May;290(9):2263-2278. doi: 10.1111/febs.16355. Epub 2022 Jan 25. FEBS J. 2023. PMID: 35032346 Review.
Cited by
-
The immune signaling pathways of Manduca sexta.Insect Biochem Mol Biol. 2015 Jul;62:64-74. doi: 10.1016/j.ibmb.2015.03.006. Epub 2015 Apr 7. Insect Biochem Mol Biol. 2015. PMID: 25858029 Free PMC article.
-
Changes in composition and levels of hemolymph proteins during metamorphosis of Manduca sexta.Insect Biochem Mol Biol. 2020 Dec;127:103489. doi: 10.1016/j.ibmb.2020.103489. Epub 2020 Oct 20. Insect Biochem Mol Biol. 2020. PMID: 33096211 Free PMC article.
-
Factor B Is the Second Lipopolysaccharide-binding Protease Zymogen in the Horseshoe Crab Coagulation Cascade.J Biol Chem. 2015 Jul 31;290(31):19379-86. doi: 10.1074/jbc.M115.653196. Epub 2015 Jun 24. J Biol Chem. 2015. PMID: 26109069 Free PMC article.
-
Building a platform for predicting functions of serine protease-related proteins in Drosophila melanogaster and other insects.Insect Biochem Mol Biol. 2018 Dec;103:53-69. doi: 10.1016/j.ibmb.2018.10.006. Epub 2018 Oct 24. Insect Biochem Mol Biol. 2018. PMID: 30367934 Free PMC article.
-
Identification of immunity-related genes distinctly regulated by Manduca sexta Spӓtzle-1/2 and Escherichia coli peptidoglycan.Insect Biochem Mol Biol. 2024 May;168:104108. doi: 10.1016/j.ibmb.2024.104108. Epub 2024 Mar 27. Insect Biochem Mol Biol. 2024. PMID: 38552808 Free PMC article.
References
-
- Barillas-Mury C. CLIP proteases and Plasmodium melanization in Anopheles gambiae. Trends Parasitol. 2007;23:297–299. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

