Protein neddylation: beyond cullin-RING ligases
- PMID: 25531226
- PMCID: PMC5131867
- DOI: 10.1038/nrm3919
Protein neddylation: beyond cullin-RING ligases
Abstract
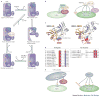
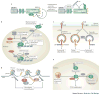
NEDD8 (neural precursor cell expressed developmentally downregulated protein 8) is a ubiquitin-like protein that activates the largest ubiquitin E3 ligase family, the cullin-RING ligases. Many non-cullin neddylation targets have been proposed in recent years. However, overexpression of exogenous NEDD8 can trigger NEDD8 conjugation through the ubiquitylation machinery, which makes validating potential NEDD8 targets challenging. Here, we re-evaluate studies of non-cullin targets of NEDD8 in light of the current understanding of the neddylation pathway, and suggest criteria for identifying genuine neddylation substrates under homeostatic conditions. We describe the biological processes that might be regulated by non-cullin neddylation, and the utility of neddylation inhibitors for research and as potential therapies. Understanding the biological significance of non-cullin neddylation is an exciting research prospect primed to reveal fundamental insights.
Figures

References
-
- van der Veen AG, Ploegh HL. Ubiquitin-like proteins. Annu Rev Biochem. 2012;81:323–357. - PubMed
-
- Schreiber A, Peter M. Substrate recognition in selective autophagy and the ubiquitin-proteasome system. Biochim Biophys Acta. 2014;1843:163–181. - PubMed
-
- Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

