PAX3 and ETS1 synergistically activate MET expression in melanoma cells
- PMID: 25531327
- PMCID: PMC4476961
- DOI: 10.1038/onc.2014.420
PAX3 and ETS1 synergistically activate MET expression in melanoma cells
Abstract
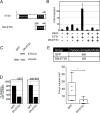
Melanoma is a highly aggressive disease that is difficult to treat owing to rapid tumor growth, apoptotic resistance and high metastatic potential. The MET proto-oncogene (MET) tyrosine kinase receptor promotes many of these cellular processes, but while MET is often overexpressed in melanoma, the mechanism driving this overexpression is unknown. As the MET gene is rarely mutated or amplified in melanoma, MET overexpression may be driven to increased activation through promoter elements. In this report, we find that transcription factors PAX3 and ETS1 directly interact to synergistically activate MET expression. Inhibition of PAX3 and ETS1 expression in melanoma cells leads to a significant reduction of MET receptor levels. The 300-bp 5' proximal MET promoter contains a PAX3 response element and two ETS1 consensus motifs. Although ETS1 can moderately activate both of these sites without cofactors, robust MET promoter activation of the first site is PAX dependent and requires the presence of PAX3, whereas the second site is PAX independent. The induction of MET by ETS1 via this second site is enhanced by hepatocyte growth factor-dependent ETS1 activation, thereby MET indirectly promotes its own expression. We further find that expression of a dominant-negative ETS1 reduces the ability of melanoma cells to grow both in culture and in vivo. Thus, we discover a pathway where ETS1 advances melanoma through the expression of MET via PAX-dependent and -independent mechanisms.
Conflict of interest statement
Figures




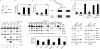
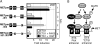
References
-
- Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. - PubMed
-
- Keehn CA, Smoller BR, Morgan MB. Expression of the ets-1 proto-oncogene in melanocytic lesions. Mod Pathol. 2003;16(8):772–7. - PubMed
-
- Tajima A, Miyamoto Y, Kadowaki H, Hayashi M. Mouse integrin alphav promoter is regulated by transcriptional factors Ets and Sp1 in melanoma cells. Biochim Biophys Acta. 2000;1492(2–3):377–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

