Integrating phosphoproteome and transcriptome reveals new determinants of macrophage multinucleation
- PMID: 25532521
- PMCID: PMC4349971
- DOI: 10.1074/mcp.M114.043836
Integrating phosphoproteome and transcriptome reveals new determinants of macrophage multinucleation
Abstract
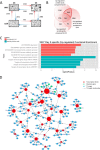
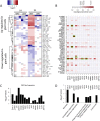
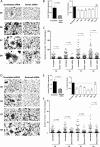
Macrophage multinucleation (MM) is essential for various biological processes such as osteoclast-mediated bone resorption and multinucleated giant cell-associated inflammatory reactions. Here we study the molecular pathways underlying multinucleation in the rat through an integrative approach combining MS-based quantitative phosphoproteomics (LC-MS/MS) and transcriptome (high-throughput RNA-sequencing) to identify new regulators of MM. We show that a strong metabolic shift toward HIF1-mediated glycolysis occurs at transcriptomic level during MM, together with modifications in phosphorylation of over 50 proteins including several ARF GTPase activators and polyphosphate inositol phosphatases. We use shortest-path analysis to link differential phosphorylation with the transcriptomic reprogramming of macrophages and identify LRRFIP1, SMARCA4, and DNMT1 as novel regulators of MM. We experimentally validate these predictions by showing that knock-down of these latter reduce macrophage multinucleation. These results provide a new framework for the combined analysis of transcriptional and post-translational changes during macrophage multinucleation, prioritizing essential genes, and revealing the sequential events leading to the multinucleation of macrophages.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
Conflict of interest: The authors declare that they have no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

