Fine mapping of Rcr1 and analyses of its effect on transcriptome patterns during infection by Plasmodiophora brassicae
- PMID: 25532522
- PMCID: PMC4326500
- DOI: 10.1186/1471-2164-15-1166
Fine mapping of Rcr1 and analyses of its effect on transcriptome patterns during infection by Plasmodiophora brassicae
Abstract
Background: The protist Plasmodiophora brassicae is a biotrophic soil-borne pathogen that causes clubroot on Brassica crops worldwide. Clubroot disease is a serious threat to the 8 M ha of canola (Brassica napus) grown annually in western Canada. While host resistance is the key to clubroot management, sources of resistance are limited.
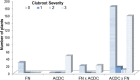
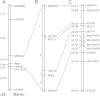
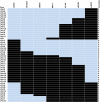
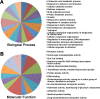
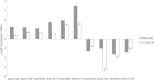
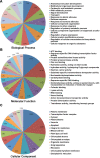
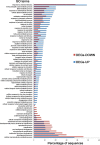
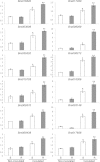
Results: To identify new sources of clubroot resistance (CR), we fine mapped a CR gene (Rcr1) from B. rapa ssp. chinensis to the region between 24.26 Mb and 24.50 Mb on the linkage group A03, with several closely linked markers identified. Transcriptome analysis was conducted using RNA sequencing on a segregating F1 population inoculated with P. brassicae, with 2,212 differentially expressed genes (DEGs) identified between plants carrying and not carrying Rcr1. Functional annotation of these DEGs showed that several defense-related biological processes, including signaling and metabolism of jasmonate and ethylene, defensive deposition of callose and biosynthesis of indole-containing compounds, were up-regulated significantly in plants carrying Rcr1 while genes involved in salicylic acid metabolic and signaling pathways were generally not elevated. Several DEGs involved in metabolism potentially related to clubroot symptom development, including auxin biosynthesis and cell growth/development, showed significantly lower expression in plants carrying Rcr1.
Conclusion: The CR gene Rcr1 and closely linked markers will be highly useful for breeding new resistant canola cultivars. The identification of DEGs between inoculated plants carrying and not carrying Rcr1 is an important step towards understanding of specific metabolic/signaling pathways in clubroot resistance mediated by Rcr1. This information may help judicious use of CR genes with complementary resistance mechanisms for durable clubroot resistance.
Figures


References
-
- Dixon GR. The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J Plant Growth Regul. 2009;28(3):194–202. doi: 10.1007/s00344-009-9090-y. - DOI
-
- Peng G, Lahlali R, Hwang SF, Pageau D, Hynes RK, McDonald MR, Gossen BD, Strelkov SE. Crop rotation, cultivar resistance, and fungicides/biofungicides for managing clubroot (Plasmodiophora brassicae) on canola. Can J Plant Pathol. 2014;36(Suppl. 1):99–112. doi: 10.1080/07060661.2013.860398. - DOI
-
- Wallenhammar AC. Prevalence of Plasmodiophora brassicae in a spring oilseed rape growing area in central Sweden and factors influencing soil infestation levels. Plant Pathol. 1996;45(4):710–719. doi: 10.1046/j.1365-3059.1996.d01-173.x. - DOI
-
- Tsusihima S, Murakami H, Akimoto T, Katahira M, Kuroyanagi Y, Shishido Y. A practical estimating method of the dose response curve between inoculum density of Plasmodiophora brassicae and the disease severity for long-term IPM strategies. JARQ. 2010;44(4):383–390. doi: 10.6090/jarq.44.383. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

