Mechanisms of virus assembly
- PMID: 25532951
- PMCID: PMC4382372
- DOI: 10.1146/annurev-physchem-040214-121637
Mechanisms of virus assembly
Abstract
Viruses are nanoscale entities containing a nucleic acid genome encased in a protein shell called a capsid and in some cases are surrounded by a lipid bilayer membrane. This review summarizes the physics that govern the processes by which capsids assemble within their host cells and in vitro. We describe the thermodynamics and kinetics for the assembly of protein subunits into icosahedral capsid shells and how these are modified in cases in which the capsid assembles around a nucleic acid or on a lipid bilayer. We present experimental and theoretical techniques used to characterize capsid assembly, and we highlight aspects of virus assembly that are likely to receive significant attention in the near future.
Keywords: RNA packaging; capsid; kinetics; membrane; simulation; thermodynamics.
Figures
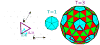

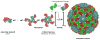

 symbols) is compared to actual charge ratios (
symbols) is compared to actual charge ratios (
 symbols) for several viruses. Predicted optimal charge ratios in the absence of base-pairing are also shown (
symbols) for several viruses. Predicted optimal charge ratios in the absence of base-pairing are also shown (
 symbols). The thermodynamic optimum charge ratio is defined as the NA length which minimizes the free energy for encapsidating the genome divided by the positive capsid charge.
symbols). The thermodynamic optimum charge ratio is defined as the NA length which minimizes the free energy for encapsidating the genome divided by the positive capsid charge.


References
-
- Ban N, McPherson A. The structure of satellite panicum mosaic virus at 1.9 Å resolution. Nat Struct Mol Biol. 1995;2:882–890. - PubMed
-
- Philippe N, Legendre M, Doutre G, Coute Y, Poirot O, et al. Pandoraviruses: Amoeba Viruses with Genomes Up to 2.5 Mb Reaching That of Parasitic Eukaryotes. Science. 2013;341:281–286. - PubMed
-
- Mateu MG. Assembly, stability and dynamics of virus capsids. Arch Biochem Biophys. 2013;531:65–79. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

