Noncovalent interactions with SUMO and ubiquitin orchestrate distinct functions of the SLX4 complex in genome maintenance
- PMID: 25533185
- PMCID: PMC4289429
- DOI: 10.1016/j.molcel.2014.11.015
Noncovalent interactions with SUMO and ubiquitin orchestrate distinct functions of the SLX4 complex in genome maintenance
Abstract
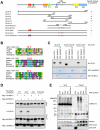
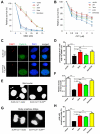
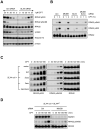
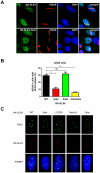
SLX4, a coordinator of multiple DNA structure-specific endonucleases, is important for several DNA repair pathways. Noncovalent interactions of SLX4 with ubiquitin are required for localizing SLX4 to DNA interstrand crosslinks (ICLs), yet how SLX4 is targeted to other functional contexts remains unclear. Here, we show that SLX4 binds SUMO-2/3 chains via SUMO-interacting motifs (SIMs). The SIMs of SLX4 are dispensable for ICL repair but important for processing CPT-induced replication intermediates, suppressing fragile site instability, and localizing SLX4 to ALT telomeres. The localization of SLX4 to laser-induced DNA damage also requires the SIMs, as well as DNA end resection, UBC9, and MDC1. Furthermore, the SUMO binding of SLX4 enhances its interaction with specific DNA-damage sensors or telomere-binding proteins, including RPA, MRE11-RAD50-NBS1, and TRF2. Thus, the interactions of SLX4 with SUMO and ubiquitin increase its affinity for factors recognizing different DNA lesions or telomeres, helping to direct the SLX4 complex in distinct functional contexts.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


Comment in
-
SLX4: not SIMply a nuclease scaffold?Mol Cell. 2015 Jan 8;57(1):3-5. doi: 10.1016/j.molcel.2014.12.032. Mol Cell. 2015. PMID: 25574947
References
-
- Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, Shirakawa M. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature. 2005;435:979–982. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

