Accessing Data Resources in the Mouse Phenome Database for Genetic Analysis of Murine Life Span and Health Span
- PMID: 25533306
- PMCID: PMC4707687
- DOI: 10.1093/gerona/glu223
Accessing Data Resources in the Mouse Phenome Database for Genetic Analysis of Murine Life Span and Health Span
Abstract
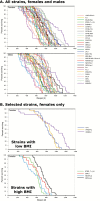
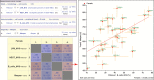
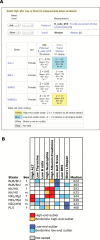
Understanding the source of genetic variation in aging and using this variation to define the molecular mechanisms of healthy aging require deep and broad quantification of a host of physiological, morphological, and behavioral endpoints. The murine model is a powerful system in which to understand the relations across age-related phenotypes and to identify research models with variation in life span and health span. The Jackson Laboratory Nathan Shock Center of Excellence in the Basic Biology of Aging has performed broad characterization of aging in genetically diverse laboratory mice and has placed these data, along with data from several other major aging initiatives, into the interactive Mouse Phenome Database. The data may be accessed and analyzed by researchers interested in finding mouse models for specific aging processes, age-related health and disease states, and for genetic analysis of aging variation and trait covariation. We expect that by placing these data in the hands of the aging community that there will be (a) accelerated genetic analyses of aging processes, (b) discovery of genetic loci regulating life span, (c) identification of compelling correlations between life span and susceptibility for age-related disorders, and (d) discovery of concordant genomic loci influencing life span and aging phenotypes between mouse and humans.
Keywords: Database; Functional genomics; Mouse; Phenotype.
© The Author 2014. Published by Oxford University Press on behalf of The Gerontological Society of America.
Figures


References
-
- Churchill GA, Airey DC, Allayee H, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36:1133–1137. doi:10.1038/ng1104-1133 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

