The FBXL10/KDM2B scaffolding protein associates with novel polycomb repressive complex-1 to regulate adipogenesis
- PMID: 25533466
- PMCID: PMC4326826
- DOI: 10.1074/jbc.M114.626929
The FBXL10/KDM2B scaffolding protein associates with novel polycomb repressive complex-1 to regulate adipogenesis
Abstract
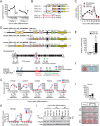
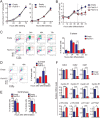
Polycomb repressive complex 1 (PRC1) plays an essential role in the epigenetic repression of gene expression during development and cellular differentiation via multiple effector mechanisms, including ubiquitination of H2A and chromatin compaction. However, whether it regulates the stepwise progression of adipogenesis is unknown. Here, we show that FBXL10/KDM2B is an anti-adipogenic factor that is up-regulated during the early phase of 3T3-L1 preadipocyte differentiation and in adipose tissue in a diet-induced model of obesity. Interestingly, inhibition of adipogenesis does not require the JmjC demethylase domain of FBXL10, but it does require the F-box and leucine-rich repeat domains, which we show recruit a noncanonical polycomb repressive complex 1 (PRC1) containing RING1B, SKP1, PCGF1, and BCOR. Knockdown of either RING1B or SKP1 prevented FBXL10-mediated repression of 3T3-L1 preadipocyte differentiation indicating that PRC1 formation mediates the inhibitory effect of FBXL10 on adipogenesis. Using ChIP-seq, we show that FBXL10 recruits RING1B to key specific genomic loci surrounding the key cell cycle and the adipogenic genes Cdk1, Uhrf1, Pparg1, and Pparg2 to repress adipogenesis. These results suggest that FBXL10 represses adipogenesis by targeting a noncanonical PRC1 complex to repress key genes (e.g. Pparg) that control conversion of pluripotent cells into the adipogenic lineage.
Keywords: 3T3-L1 cells; Adipocyte; Adipogenesis; Cell Differentiation; FBXL10/KDM2B/JHDM1B; Mitotic Clonal Expansion (MCE); Peroxisome Proliferator-activated Receptor (PPAR); Polycomb; Polycomb Repressive Complex 1 (PRC1); RING1B.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Rosen E. D., MacDougald O. A. (2006) Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 7, 885–896 - PubMed
-
- Wu Z., Rosen E. D., Brun R., Hauser S., Adelmant G., Troy A. E., McKeon C., Darlington G. J., Spiegelman B. M. (1999) Cross-regulation of C/EBPα and PPARγ controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol. Cell 3, 151–158 - PubMed
-
- MacDougald O. A., Lane M. D. (1995) Transcriptional regulation of gene expression during adipocyte differentiation. Annu. Rev. Biochem. 64, 345–373 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

