Long-term culture of genome-stable bipotent stem cells from adult human liver
- PMID: 25533785
- PMCID: PMC4313365
- DOI: 10.1016/j.cell.2014.11.050
Long-term culture of genome-stable bipotent stem cells from adult human liver
Abstract
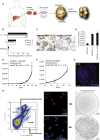
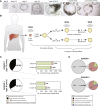
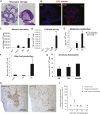
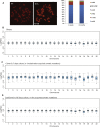
Despite the enormous replication potential of the human liver, there are currently no culture systems available that sustain hepatocyte replication and/or function in vitro. We have shown previously that single mouse Lgr5+ liver stem cells can be expanded as epithelial organoids in vitro and can be differentiated into functional hepatocytes in vitro and in vivo. We now describe conditions allowing long-term expansion of adult bile duct-derived bipotent progenitor cells from human liver. The expanded cells are highly stable at the chromosome and structural level, while single base changes occur at very low rates. The cells can readily be converted into functional hepatocytes in vitro and upon transplantation in vivo. Organoids from α1-antitrypsin deficiency and Alagille syndrome patients mirror the in vivo pathology. Clonal long-term expansion of primary adult liver stem cells opens up experimental avenues for disease modeling, toxicology studies, regenerative medicine, and gene therapy.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures










Comment in
-
Organoid cultures boost human liver cell expansion.Hepatology. 2015 Nov;62(5):1635-7. doi: 10.1002/hep.27970. Epub 2015 Jul 31. Hepatology. 2015. PMID: 26146934 No abstract available.
References
-
- Baker D.E., Harrison N.J., Maltby E., Smith K., Moore H.D., Shaw P.J., Heath P.R., Holden H., Andrews P.W. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat. Biotechnol. 2007;25:207–215. - PubMed
-
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. - PubMed
-
- Barker N., Huch M., Kujala P., van de Wetering M., Snippert H.J., van Es J.H., Sato T., Stange D.E., Begthel H., van den Born M. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. - PubMed
Supplemental References
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

