Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds
- PMID: 25533786
- PMCID: PMC4297522
- DOI: 10.1016/j.cell.2014.11.052
Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds
Abstract
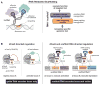
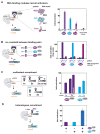
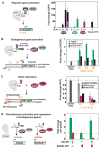
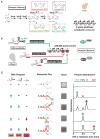
Eukaryotic cells execute complex transcriptional programs in which specific loci throughout the genome are regulated in distinct ways by targeted regulatory assemblies. We have applied this principle to generate synthetic CRISPR-based transcriptional programs in yeast and human cells. By extending guide RNAs to include effector protein recruitment sites, we construct modular scaffold RNAs that encode both target locus and regulatory action. Sets of scaffold RNAs can be used to generate synthetic multigene transcriptional programs in which some genes are activated and others are repressed. We apply this approach to flexibly redirect flux through a complex branched metabolic pathway in yeast. Moreover, these programs can be executed by inducing expression of the dCas9 protein, which acts as a single master regulatory control point. CRISPR-associated RNA scaffolds provide a powerful way to construct synthetic gene expression programs for a wide range of applications, including rewiring cell fates or engineering metabolic pathways.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Technology: multi-tasking CRISPR RNA scaffolds.Nat Rev Genet. 2015 Feb;16(2):70. doi: 10.1038/nrg3898. Epub 2015 Jan 2. Nat Rev Genet. 2015. PMID: 25554357 No abstract available.
-
Complex regulatory control with CRISPR.Nat Methods. 2015 Mar;12(3):172. doi: 10.1038/nmeth.3308. Nat Methods. 2015. PMID: 25879101 No abstract available.
-
Toward Whole-Transcriptome Editing with CRISPR-Cas9.Mol Cell. 2015 May 21;58(4):560-2. doi: 10.1016/j.molcel.2015.05.016. Mol Cell. 2015. PMID: 26000839
References
-
- Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314:1565–1568. - PubMed
-
- Andronescu M, Fejes AP, Hutter F, Hoos HH, Condon A. A new algorithm for RNA secondary structure design. J Mol Biol. 2004;336:607–624. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

