An Arabidopsis gene regulatory network for secondary cell wall synthesis
- PMID: 25533953
- PMCID: PMC4333722
- DOI: 10.1038/nature14099
An Arabidopsis gene regulatory network for secondary cell wall synthesis
Abstract
The plant cell wall is an important factor for determining cell shape, function and response to the environment. Secondary cell walls, such as those found in xylem, are composed of cellulose, hemicelluloses and lignin and account for the bulk of plant biomass. The coordination between transcriptional regulation of synthesis for each polymer is complex and vital to cell function. A regulatory hierarchy of developmental switches has been proposed, although the full complement of regulators remains unknown. Here we present a protein-DNA network between Arabidopsis thaliana transcription factors and secondary cell wall metabolic genes with gene expression regulated by a series of feed-forward loops. This model allowed us to develop and validate new hypotheses about secondary wall gene regulation under abiotic stress. Distinct stresses are able to perturb targeted genes to potentially promote functional adaptation. These interactions will serve as a foundation for understanding the regulation of a complex, integral plant component.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

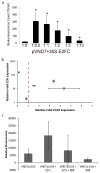



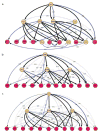
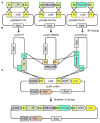



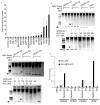
Comment in
-
Plant biology: Seeing the wood and the trees.Nature. 2015 Jan 29;517(7536):558-9. doi: 10.1038/nature14085. Epub 2014 Dec 31. Nature. 2015. PMID: 25533950 No abstract available.
References
-
- Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

