Marker profile for the evaluation of human umbilical artery smooth muscle cell quality obtained by different isolation and culture methods
- PMID: 25535117
- PMCID: PMC4960121
- DOI: 10.1007/s10616-014-9822-0
Marker profile for the evaluation of human umbilical artery smooth muscle cell quality obtained by different isolation and culture methods
Abstract
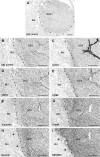
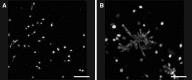
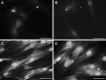
Even though umbilical cord arteries are a common source of vascular smooth muscle cells, the lack of reliable marker profiles have not facilitated the isolation of human umbilical artery smooth muscle cells (HUASMC). For accurate characterization of HUASMC and cells in their environment, the expression of smooth muscle and mesenchymal markers was analyzed in umbilical cord tissue sections. The resulting marker profile was then used to evaluate the quality of HUASMC isolation and culture methods. HUASMC and perivascular-Wharton's jelly stromal cells (pv-WJSC) showed positive staining for α-smooth muscle actin (α-SMA), smooth muscle myosin heavy chain (SM-MHC), desmin, vimentin and CD90. Anti-CD10 stained only pv-WJSC. Consequently, HUASMC could be characterized as α-SMA+ , SM-MHC+ , CD10- cells, which are additionally negative for endothelial markers (CD31 and CD34). Enzymatic isolation provided primary HUASMC batches with 90-99 % purity, yet, under standard culture conditions, contaminant CD10+ cells rapidly constituted more than 80 % of the total cell population. Contamination was mainly due to the poor adhesion of HUASMC to cell culture plates, regardless of the different protein coatings (fibronectin, collagen I or gelatin). HUASMC showed strong attachment and long-term viability only in 3D matrices. The explant isolation method achieved cultures with only 13-40 % purity with considerable contamination by CD10+ cells. CD10+ cells showed spindle-like morphology and up-regulated expression of α-SMA and SM-MHC upon culture in smooth muscle differentiation medium. Considering the high contamination risk of HUASMC cultures by CD10+ neighboring cells and their phenotypic similarities, precise characterization is mandatory to avoid misleading results.
Keywords: CD10; Characterization; Human umbilical artery smooth muscle cells (HUASMC); Isolation; Markers; Wharton’s jelly stromal cells.
Figures

Similar articles
-
[Biological characteristics of human umbilical artery smooth muscle cells cultured in vitro and the preestablishment of immortalized cell line].Sichuan Da Xue Xue Bao Yi Xue Ban. 2003 Apr;34(2):189-92. Sichuan Da Xue Xue Bao Yi Xue Ban. 2003. PMID: 12947685 Chinese.
-
[Effects of nuclear factor-kappaB decoy oligodeoxynucleotide on the function of human umbilical artery smooth muscle cells induced by umbilical sera in preeclampsia].Zhonghua Fu Chan Ke Za Zhi. 2007 Feb;42(2):87-91. Zhonghua Fu Chan Ke Za Zhi. 2007. PMID: 17442180 Chinese.
-
Isolation and culture of human umbilical artery smooth muscle cells expressing functional calcium channels.In Vitro Cell Dev Biol Anim. 2009 Mar-Apr;45(3-4):175-84. doi: 10.1007/s11626-008-9161-6. Epub 2009 Jan 1. In Vitro Cell Dev Biol Anim. 2009. PMID: 19118440
-
Use of histone deacetylase 8 (HDAC8), a new marker of smooth muscle differentiation, in the classification of mesenchymal tumors of the uterus.Am J Surg Pathol. 2006 Mar;30(3):319-27. doi: 10.1097/01.pas.0000188029.63706.31. Am J Surg Pathol. 2006. PMID: 16538051
-
Perspectives of employing mesenchymal stem cells from the Wharton's jelly of the umbilical cord for peripheral nerve repair.Int Rev Neurobiol. 2013;108:79-120. doi: 10.1016/B978-0-12-410499-0.00004-6. Int Rev Neurobiol. 2013. PMID: 24083432 Review.
Cited by
-
miR-195 inhibits macrophages pro-inflammatory profile and impacts the crosstalk with smooth muscle cells.PLoS One. 2017 Nov 22;12(11):e0188530. doi: 10.1371/journal.pone.0188530. eCollection 2017. PLoS One. 2017. PMID: 29166412 Free PMC article.
-
Perinatal Derivatives: Where Do We Stand? A Roadmap of the Human Placenta and Consensus for Tissue and Cell Nomenclature.Front Bioeng Biotechnol. 2020 Dec 17;8:610544. doi: 10.3389/fbioe.2020.610544. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 33392174 Free PMC article. Review.
-
Placental Tissues as Biomaterials in Regenerative Medicine.Biomed Res Int. 2022 Apr 21;2022:6751456. doi: 10.1155/2022/6751456. eCollection 2022. Biomed Res Int. 2022. PMID: 35496035 Free PMC article. Review.
-
Glucose degradation products in peritoneal dialysis solution impair angiogenesis by dysregulating angiogenetic factors in endothelial and vascular smooth muscle cells.Clin Exp Nephrol. 2022 Dec;26(12):1160-1169. doi: 10.1007/s10157-022-02272-3. Epub 2022 Sep 7. Clin Exp Nephrol. 2022. PMID: 36070106
References
-
- Corrao S, La Rocca G, Lo IM, Corsello T, Farina F, Anzalone R. Umbilical cord revisited: from Wharton’s jelly myofibroblasts to mesenchymal stem cells. Histol Histopathol. 2013;28:1235–1244. - PubMed
-
- Farias VA, Linares-Fernandez JL, Penalver JL, Paya Colmenero JA, Ferron GO, Duran EL, Fernandez RM, Olivares EG, O’Valle F, Puertas A, Oliver FJ, Ruiz de Almodovar JM. Human umbilical cord stromal stem cell express CD10 and exert contractile properties. Placenta. 2011;32:86–95. doi: 10.1016/j.placenta.2010.11.003. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

