A novel DNA-binding protein plays an important role in Helicobacter pylori stress tolerance and survival in the host
- PMID: 25535274
- PMCID: PMC4325109
- DOI: 10.1128/JB.02489-14
A novel DNA-binding protein plays an important role in Helicobacter pylori stress tolerance and survival in the host
Abstract
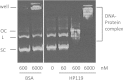
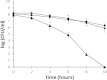
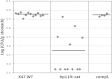
The gastric pathogen Helicobacter pylori must combat chronic acid and oxidative stress. It does so via many mechanisms, including macromolecule repair and gene regulation. Mitomycin C-sensitive clones from a transposon mutagenesis library were screened. One sensitive strain contained the insertion element at the locus of hp119, a hypothetical gene. No homologous gene exists in any (non-H. pylori) organism. Nevertheless, the predicted protein has some features characteristic of histone-like proteins, and we showed that purified HP119 protein is a DNA-binding protein. A Δhp119 strain was markedly more sensitive (viability loss) to acid or to air exposure, and these phenotypes were restored to wild-type (WT) attributes upon complementation of the mutant with the wild-type version of hp119 at a separate chromosomal locus. The mutant strain was approximately 10-fold more sensitive to macrophage-mediated killing than the parent or the complemented strain. Of 12 mice inoculated with the wild type, all contained H. pylori, whereas 5 of 12 mice contained the mutant strain; the mean colonization numbers were 158-fold less for the mutant strain. A proteomic (two-dimensional PAGE with mass spectrometric analysis) comparison between the Δhp119 mutant and the WT strain under oxidative stress conditions revealed a number of important antioxidant protein differences; SodB, Tpx, TrxR, and NapA, as well as the peptidoglycan deacetylase PgdA, were significantly less expressed in the Δhp119 mutant than in the WT strain. This study identified HP119 as a putative histone-like DNA-binding protein and showed that it plays an important role in Helicobacter pylori stress tolerance and survival in the host.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
References
-
- McGee DJ, Mobley HL. 1999. Mechanisms of Helicobacter pylori infection: bacterial factors. Curr Top Microbiol Immunol 241:155–180. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

