Rapid evolution of the env gene leader sequence in cats naturally infected with feline immunodeficiency virus
- PMID: 25535323
- PMCID: PMC4361796
- DOI: 10.1099/vir.0.000035
Rapid evolution of the env gene leader sequence in cats naturally infected with feline immunodeficiency virus
Abstract
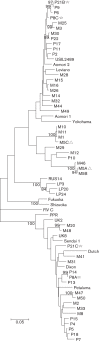
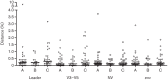
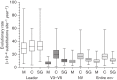
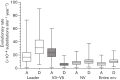
Analysing the evolution of feline immunodeficiency virus (FIV) at the intra-host level is important in order to address whether the diversity and composition of viral quasispecies affect disease progression. We examined the intra-host diversity and the evolutionary rates of the entire env and structural fragments of the env sequences obtained from sequential blood samples in 43 naturally infected domestic cats that displayed different clinical outcomes. We observed in the majority of cats that FIV env showed very low levels of intra-host diversity. We estimated that env evolved at a rate of 1.16×10(-3) substitutions per site per year and demonstrated that recombinant sequences evolved faster than non-recombinant sequences. It was evident that the V3-V5 fragment of FIV env displayed higher evolutionary rates in healthy cats than in those with terminal illness. Our study provided the first evidence that the leader sequence of env, rather than the V3-V5 sequence, had the highest intra-host diversity and the highest evolutionary rate of all env fragments, consistent with this region being under a strong selective pressure for genetic variation. Overall, FIV env displayed relatively low intra-host diversity and evolved slowly in naturally infected cats. The maximum evolutionary rate was observed in the leader sequence of env. Although genetic stability is not necessarily a prerequisite for clinical stability, the higher genetic stability of FIV compared with human immunodeficiency virus might explain why many naturally infected cats do not progress rapidly to AIDS.
© 2015 The Authors.
Figures
References
-
- Bęczkowski P. M. (2013). Virus evolution in the progression of natural feline immunodeficiency virus infection. PhD thesis, Centre for Virus Research, University of Glasgow, UK.
-
- Bęczkowski P. M., Techakriengkrai N., Logan N., McMonagle E., Litster A., Willett B. J., Hosie M. J. (2014b). Emergence of CD134 cysteine-rich domain 2 (CRD2)-independent strains of feline immunodeficiency virus (FIV) is associated with disease progression in naturally infected cats. Retrovirology 11, 95. 10.1186/s12977-014-0095-7 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

