Patterns of accumulation of miRNAs encoded by herpes simplex virus during productive infection, latency, and on reactivation
- PMID: 25535379
- PMCID: PMC4291656
- DOI: 10.1073/pnas.1422657112
Patterns of accumulation of miRNAs encoded by herpes simplex virus during productive infection, latency, and on reactivation
Erratum in
-
Correction for Du et al., Patterns of accumulation of miRNAs encoded by herpes simplex virus during productive infection, latency, and on reactivation.Proc Natl Acad Sci U S A. 2016 Jan 5;113(1):E102. doi: 10.1073/pnas.1524373113. Epub 2015 Dec 28. Proc Natl Acad Sci U S A. 2016. PMID: 26711985 Free PMC article. No abstract available.
Abstract
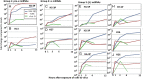
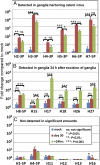
The key events in herpes simplex virus (HSV) infections are (i) replication at a portal of entry into the body modeled by infection of cultured cells; (ii) establishment of a latent state characterized by a sole latency-associated transcript and microRNAs (miRNAs) modeled in murine peripheral ganglia 30 d after inoculation; and (iii) reactivation from the latent state modeled by excision and incubation of ganglia in medium containing anti-NGF antibody for a timespan of a single viral replicative cycle. In this report, we examine the pattern of synthesis and accumulation of 18 HSV-1 miRNAs in the three models. We report the following: (i) H2-3P, H3-3P, H4-3P, H5-3P, H6-3P, and H7-5P accumulated in ganglia harboring latent virus. All but H4-3P were readily detected in productively infected cells, and most likely they originate from three transcriptional units. (ii) H8-5P, H15, H17, H18, H26, and H27 accumulated during reactivation. Of this group, only H26 and H27 could be detected in productively infected cells. (iii) Of the 18 we have examined, only 10 miRNAs were found to accumulate above background levels in productively infected cells. The disparity in the accumulation of miRNAs in cell culture and during reactivation may reflect differences in the patterns of regulation of viral gene expression during productive infection and during reactivation from the latent state.
Keywords: alpha genes; beta/gamma genes; regulation of gene expression; transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Deletion of Herpes Simplex Virus 1 MicroRNAs miR-H1 and miR-H6 Impairs Reactivation.J Virol. 2020 Jul 16;94(15):e00639-20. doi: 10.1128/JVI.00639-20. Print 2020 Jul 16. J Virol. 2020. PMID: 32295910 Free PMC article.
-
Effect of Loss-of-function of the Herpes Simplex Virus-1 microRNA H6-5p on Virus Replication.Virol Sin. 2019 Aug;34(4):386-396. doi: 10.1007/s12250-019-00111-6. Epub 2019 Apr 24. Virol Sin. 2019. PMID: 31020575 Free PMC article.
-
Herpes Simplex Virus 1 Lytic Infection Blocks MicroRNA (miRNA) Biogenesis at the Stage of Nuclear Export of Pre-miRNAs.mBio. 2019 Feb 12;10(1):e02856-18. doi: 10.1128/mBio.02856-18. mBio. 2019. PMID: 30755517 Free PMC article.
-
Use of laser capture microdissection together with in situ hybridization and real-time PCR to study distribution of latent herpes simplex virus genomes in mouse trigeminal ganglia.Methods Mol Biol. 2005;293:285-93. doi: 10.1385/1-59259-853-6:285. Methods Mol Biol. 2005. PMID: 16028427 Review.
-
Restarting Lytic Gene Transcription at the Onset of Herpes Simplex Virus Reactivation.J Virol. 2017 Jan 3;91(2):e01419-16. doi: 10.1128/JVI.01419-16. Print 2017 Jan 15. J Virol. 2017. PMID: 27807236 Free PMC article. Review.
Cited by
-
An in vitro model of latency and reactivation of varicella zoster virus in human stem cell-derived neurons.PLoS Pathog. 2015 Jun 4;11(6):e1004885. doi: 10.1371/journal.ppat.1004885. eCollection 2015 Jun. PLoS Pathog. 2015. PMID: 26042814 Free PMC article.
-
Induction of varicella zoster virus DNA replication in dissociated human trigeminal ganglia.J Neurovirol. 2017 Feb;23(1):152-157. doi: 10.1007/s13365-016-0480-1. Epub 2016 Sep 28. J Neurovirol. 2017. PMID: 27683235 Free PMC article.
-
Human herpesvirus-encoded MicroRNA in host-pathogen interaction.Adv Biol Regul. 2021 Dec;82:100829. doi: 10.1016/j.jbior.2021.100829. Epub 2021 Sep 16. Adv Biol Regul. 2021. PMID: 34560402 Free PMC article. Review.
-
Viral MicroRNAs in Herpes Simplex Virus 1 Pathobiology.Curr Pharm Des. 2024;30(9):649-665. doi: 10.2174/0113816128286469240129100313. Curr Pharm Des. 2024. PMID: 38347772 Review.
-
The Role of microRNAs in the Pathogenesis of Herpesvirus Infection.Viruses. 2016 Jun 2;8(6):156. doi: 10.3390/v8060156. Viruses. 2016. PMID: 27271654 Free PMC article. Review.
References
-
- Roizman B, Knipe DM, Whitley RJ. Herpes simplex viruses. In: Knipe DM, et al., editors. Fields' Virology. 6th Ed. Wolters Kluwer/Lippincott-Williams and Wilkins; New York: 2013. pp. 1823–1897.
-
- Stevens JG, Wagner EK, Devi-Rao GB, Cook ML, Feldman LT. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science. 1987;235(4792):1056–1059. - PubMed
-
- Spivack JG, Woods GM, Fraser NW. Identification of a novel latency-specific splice donor signal within the herpes simplex virus type 1 2.0-kilobase latency-associated transcript (LAT): Translation inhibition of LAT open reading frames by the intron within the 2.0-kilobase LAT. J Virol. 1991;65(12):6800–6810. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

