Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins
- PMID: 25535392
- PMCID: PMC4291625
- DOI: 10.1073/pnas.1417910112
Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins
Abstract
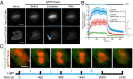
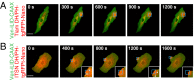
The discovery of light-inducible protein-protein interactions has allowed for the spatial and temporal control of a variety of biological processes. To be effective, a photodimerizer should have several characteristics: it should show a large change in binding affinity upon light stimulation, it should not cross-react with other molecules in the cell, and it should be easily used in a variety of organisms to recruit proteins of interest to each other. To create a switch that meets these criteria we have embedded the bacterial SsrA peptide in the C-terminal helix of a naturally occurring photoswitch, the light-oxygen-voltage 2 (LOV2) domain from Avena sativa. In the dark the SsrA peptide is sterically blocked from binding its natural binding partner, SspB. When activated with blue light, the C-terminal helix of the LOV2 domain undocks from the protein, allowing the SsrA peptide to bind SspB. Without optimization, the switch exhibited a twofold change in binding affinity for SspB with light stimulation. Here, we describe the use of computational protein design, phage display, and high-throughput binding assays to create an improved light inducible dimer (iLID) that changes its affinity for SspB by over 50-fold with light stimulation. A crystal structure of iLID shows a critical interaction between the surface of the LOV2 domain and a phenylalanine engineered to more tightly pin the SsrA peptide against the LOV2 domain in the dark. We demonstrate the functional utility of the switch through light-mediated subcellular localization in mammalian cell culture and reversible control of small GTPase signaling.
Keywords: PER-ARNT-SIM domain; Rosetta molecular modeling suite; computational library; optogenetic tool; phage display.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

