Relationships between protein-encoding gene abundance and corresponding process are commonly assumed yet rarely observed
- PMID: 25535936
- PMCID: PMC4511926
- DOI: 10.1038/ismej.2014.252
Relationships between protein-encoding gene abundance and corresponding process are commonly assumed yet rarely observed
Abstract
For any enzyme-catalyzed reaction to occur, the corresponding protein-encoding genes and transcripts are necessary prerequisites. Thus, a positive relationship between the abundance of gene or transcripts and corresponding process rates is often assumed. To test this assumption, we conducted a meta-analysis of the relationships between gene and/or transcript abundances and corresponding process rates. We identified 415 studies that quantified the abundance of genes or transcripts for enzymes involved in carbon or nitrogen cycling. However, in only 59 of these manuscripts did the authors report both gene or transcript abundance and rates of the appropriate process. We found that within studies there was a significant but weak positive relationship between gene abundance and the corresponding process. Correlations were not strengthened by accounting for habitat type, differences among genes or reaction products versus reactants, suggesting that other ecological and methodological factors may affect the strength of this relationship. Our findings highlight the need for fundamental research on the factors that control transcription, translation and enzyme function in natural systems to better link genomic and transcriptomic data to ecosystem processes.
Figures
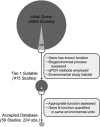
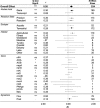
References
-
- Aloe AM, Becker BJ. Teacher verbal ability and school outcomes: where is the evidence. Educ Res. 2009;38:612–624.
-
- Brezna B, Khan AA, Cerniglia CE. Molecular characterization of dioxygenases from polycyclic aromatic hydrocarbon-degrading Mycobacterium spp. FEMS Microbiol Lett. 2003;223:177–183. - PubMed
-
- Brune A, Frenzel P, Cypionka H. Life at the oxic-anoxic interface: microbial activities and adaptations. FEMS Microbiol Rev. 2000;24:691–710. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

