Identification of molecular markers associated with Verticillium wilt resistance in alfalfa (Medicago sativa L.) using high-resolution melting
- PMID: 25536106
- PMCID: PMC4275272
- DOI: 10.1371/journal.pone.0115953
Identification of molecular markers associated with Verticillium wilt resistance in alfalfa (Medicago sativa L.) using high-resolution melting
Abstract
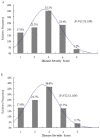
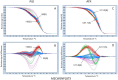
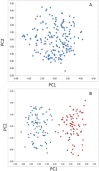
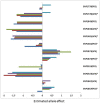
Verticillium wilt, caused by the soilborne fungus, Verticillium alfalfae, is one of the most serious diseases of alfalfa (Medicago sativa L.) worldwide. To identify loci associated with resistance to Verticillium wilt, a bulk segregant analysis was conducted in susceptible or resistant pools constructed from 13 synthetic alfalfa populations, followed by association mapping in two F1 populations consisted of 352 individuals. Simple sequence repeat (SSR) and single nucleotide polymorphism (SNP) markers were used for genotyping. Phenotyping was done by manual inoculation of the pathogen to replicated cloned plants of each individual and disease severity was scored using a standard scale. Marker-trait association was analyzed by TASSEL. Seventeen SNP markers significantly associated with Verticillium wilt resistance were identified and they were located on chromosomes 1, 2, 4, 7 and 8. SNP markers identified on chromosomes 2, 4 and 7 co-locate with regions of Verticillium wilt resistance loci reported in M. truncatula. Additional markers identified on chromosomes 1 and 8 located the regions where no Verticillium resistance locus has been reported. This study highlights the value of SNP genotyping by high resolution melting to identify the disease resistance loci in tetraploid alfalfa. With further validation, the markers identified in this study could be used for improving resistance to Verticillium wilt in alfalfa breeding programs.
Conflict of interest statement
Figures

Similar articles
-
Genotyping-by-sequencing-based genome-wide association studies on Verticillium wilt resistance in autotetraploid alfalfa (Medicago sativa L.).Mol Plant Pathol. 2017 Feb;18(2):187-194. doi: 10.1111/mpp.12389. Epub 2016 Jul 10. Mol Plant Pathol. 2017. PMID: 26933934 Free PMC article.
-
The Impact of Genotyping-by-Sequencing Pipelines on SNP Discovery and Identification of Markers Associated with Verticillium Wilt Resistance in Autotetraploid Alfalfa (Medicago sativa L.).Front Plant Sci. 2017 Feb 7;8:89. doi: 10.3389/fpls.2017.00089. eCollection 2017. Front Plant Sci. 2017. PMID: 28223988 Free PMC article.
-
Effect of Alfalfa Cultivars and Ages on the Occurrence of Verticillium Wilt Caused by Verticillium alfalfae.Plant Dis. 2022 Feb;106(2):496-503. doi: 10.1094/PDIS-04-21-0886-RE. Epub 2022 Feb 3. Plant Dis. 2022. PMID: 34420361
-
Construction of High-Density Linkage Maps and Identification of Quantitative Trait Loci Associated with Verticillium Wilt Resistance in Autotetraploid Alfalfa (Medicago sativa L.).Plant Dis. 2020 May;104(5):1439-1444. doi: 10.1094/PDIS-08-19-1718-RE. Epub 2020 Mar 9. Plant Dis. 2020. PMID: 32150504
-
A Review of Control Options and Externalities for Verticillium Wilts.Phytopathology. 2018 Feb;108(2):160-171. doi: 10.1094/PHYTO-03-17-0083-RVW. Epub 2017 Oct 16. Phytopathology. 2018. PMID: 28703041 Review.
Cited by
-
Quantitative Resistance to Verticillium Wilt in Medicago truncatula Involves Eradication of the Fungus from Roots and Is Associated with Transcriptional Responses Related to Innate Immunity.Front Plant Sci. 2016 Sep 29;7:1431. doi: 10.3389/fpls.2016.01431. eCollection 2016. Front Plant Sci. 2016. PMID: 27746789 Free PMC article.
-
Discrimination of single-point mutations in unamplified genomic DNA via Cas9 immobilized on a graphene field-effect transistor.Nat Biomed Eng. 2021 Jul;5(7):713-725. doi: 10.1038/s41551-021-00706-z. Epub 2021 Apr 5. Nat Biomed Eng. 2021. PMID: 33820980
-
Genomic Variations and Mutational Events Associated with Plant-Pathogen Interactions.Biology (Basel). 2022 Mar 10;11(3):421. doi: 10.3390/biology11030421. Biology (Basel). 2022. PMID: 35336795 Free PMC article. Review.
-
Genomics of Plant Disease Resistance in Legumes.Front Plant Sci. 2019 Oct 30;10:1345. doi: 10.3389/fpls.2019.01345. eCollection 2019. Front Plant Sci. 2019. PMID: 31749817 Free PMC article. Review.
-
Genotyping-by-sequencing-based genome-wide association studies on Verticillium wilt resistance in autotetraploid alfalfa (Medicago sativa L.).Mol Plant Pathol. 2017 Feb;18(2):187-194. doi: 10.1111/mpp.12389. Epub 2016 Jul 10. Mol Plant Pathol. 2017. PMID: 26933934 Free PMC article.
References
-
- Viands DR, Lowe CC, Bergstrom GC, Vaugh DL, Hansen JL. (1992) Association of level of resistance to Verticillium wilt with alfalfa foarge yield and stand. J Prod Agric 5:504–509.
-
- Huang HC (2003) Verticillium wilt of alfalfa: epidemiology and control strategies. Can J Plant Pathol 25:328–338.
-
- Leath KT, Erwin DC, Giffin GD (1988) Diseases and nematodes. In: Hanson AA, Barnes DK, Hill RR, editors. Alfalfa and alfalfa improvement. ASA, CSSA, SSSA, Madison, WI. pp. 621–670.
-
- Huang HC, Hanna MR, Kokko EG (1985) Mechanisms of seed contamination by Verticillium albo-atrum in alfalfa. Phytopathology 75:482–488.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

